BME103:T930 Group 17
| Home People Lab Write-Up 1 Lab Write-Up 2 Lab Write-Up 3 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
OUR TEAM
LAB 1 WRITE-UPThe Original DesignThis is an Open PCR Machine. An Open PCR Machine can rapidly duplicate DNA, or in other terms amplify it, as well as attach marker to make traits such as cancer visible. PCR stands for polymerase chain reaction. It works by heating up samples to first denature DNA and create single stranded DNA. Then it cools to allow the primer to attach and replicate the DNA. The open PCR machine starts with an initialization step where the temperature rapidly increases to 95 degrees Celsius to create a hot-start for DNA polymerization that requires heat activation. The second step is to denature the protein, where the first cycling event heats the DNA strands at a temperature of 95 degrees Celsius for 30 seconds to melt the DNA template through the disruption of hydrogen bonding between paired bases, effectively splitting the double stranded helix into two single strands of DNA. The third step is the annealing step where the temperature is rapidly lowered to around 50 degrees Celsius to allow for the annealing of primers. The polymerase then binds to the hybrid of primers with the template to begin DNA formation. Then begins the elongation step, which differs depending on the polymerase used; typically the optimum temperature is around 75 degrees Celsius. During the elongation process, DNA polymerase synthesizes a complementary new strand of anti-parallel DNA. The amount of time required for elongation differs depending on the DNA polymerase used as well as the length of the amplified DNA fragments being used. On average, DNA polymerase amplifies at a rate of one thousand bases per minute. Next is the final elongation step where the temperature is held around 75 degrees Celsius to ensure that the DNA strand is fully elongated and will generally hold for around five minutes. Finally there is an end hold temperature that keeps the reaction at a steady temperature (between four and fifteen degrees) until the amplified DNA is ready to be utilized and further studied. This is the LCD monitor and the PCB board The LCD monitor serves the purpose of informing the user of the temperature of the lid as well as the temperature of the samples. This serves as a visual verification that the program is running according to plan and follow along with the timing and temperatures involved in Open PCR experiments. This is the Heat Sink and Fan In this image the heat sink and fan have been isolated from the machine. These are used to keep the machine cool and running. When the cooling cycles begin the fan will increase speed to gradually decrease the temperature. This is the Open PCR Circuit Board In this image the circuit board has been isolated from the machine. The circuit board is used to support all of the electronic components. All of the wires connecting the fan, the LCD monitor, power supplies, and the heat sink all of which work together to ensure the functionality of the machine This is the PCR machine In this image the power supply has been isolated from the machine. The power supply provides energy for all of the components in the PCR Machine. This is the Sample Holder In this image the top has been moved aside so that the sample holder is visible. The sample holder keeps the samples in place while the PCR machine cycles. The heated lid tightens so that the inside of the lid will press against the samples without crushing them to ensure that they are heated quickly and evenly.
Experimenting With the ConnectionsWhen the LCD Monitor was unplugged from the Open PCR Circuit Board the monitor lost power. When we unplugged the white wire that connects the Open PCR Circuit Board to the Sample Holder/Heating Block, the LCD monitor incorrectly displayed the temperature. Instead of displaying the correct temperature of 25 degrees Celsius it displayed a temperature of -40 degrees Celsius.
Test RunWe first tested our PCR machine on 10/18/12. The machine malfunctioned due to lack of heat management and would not cool. It was later discovered that several internal wires were disconnected. A different machine was procured, tested, and found to work. This new machine was used for all of the experiments.
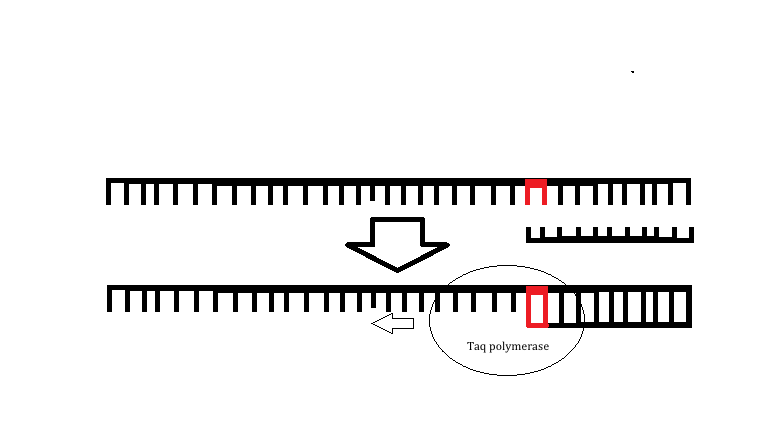
ProtocolsPolymerase Chain Reaction Part I: Description of PCR: The Polymerase Chain Reaction uses multiple heating and cooling cycles to target and amplify a specific piece of DNA in a sequence. This is useful because it allows for millions of copies of the specific DNA piece to be copied, which yields an abundant amount of samples for the person examining the DNA for disease, mutation, or etc. We used three stages for this experiment. Stage one had one cycle heating to 95 degrees Celsius for three minutes, which resulted in the DNA unwinding to a single strand. Stage 2 consisted of 35 cycles. Each cycle of stage 2 was: held at 95 degrees Celsius for 30 seconds (DNA continues to unwind), cooled to 57 degrees Celsius for 30 seconds (primer begins to bond), and re-heated to 72 degrees Celsius for 30 seconds (extension of new copy begins). Stage 3 was held at 72 degrees Celsius for 3 minutes in order to finish the extension of the DNA copying. Part II: Procedure describing amplification of patient DNA: 1. Gather all components for PCR reaction (template DNA, primers, Taq polymerase, magnesium chloride, and dNTP’s). Part III: List of all PCR components in master mix: 1. GoTaq® Colorless Master Mix, 2X Part IV: Table listing reagent and volume used:
Part V: Patient Sample Description:
Part 1: Photos of Assembly:
Part II: Assembly Procedure:
Research and DevelopmentSpecific Cancer Marker Detection - The Underlying Technology The specific DNA sequence that we are investigating is the r17879961 cancer associated sequence. This point mutation will change a thymine in a normal DNA strand to a cytosine in a mutated DNA strand. This mutation will result in an amino acid change of isoleucine to threonine when translated into an amino acid sequence. The primer designed for this single DNA mutation that causes cancer is able to bind to the template strand only if the mutation for cancer is present, which will allow for TAQ polymerase to extend the DNA. If the mutation is not present then the primer cannot bind and will therefore not be able to be extended resulting in a negative PCR reaction as amplification will not occur. The primers that we will use for this specific PCR have a sequence of AAACTCTTACACTGCATACA and CAGGACAAATTTCCTCCTAT.
Bayes's Theorem: p(A|X) = [ p(X|A)*p(A)] / [ P(X|A)*p(A) + p(X|~A)*p(~A) ] This theorem demonstrates the probability of some event happening based on a certain observation. A is the given phenomenon, while X is some observation of the phenomenon. In our case the phenomenon was either positive for cancer or negative for cancer and the observation of the phenomenon was weather it was positive or negative. Therefore, we had two equations derived from this model: PPV = TP/(TP+FP) PPV is positive prediction value
TP is true positive
FP is false positive
This gives you the probability that someone actually does have cancer.
and NPV = TN/(TN + FN) NPV is negative prediction value
TN is true negative
FN is false negative
This gives you the probability that someone does not have cancer.
Results  
Supporting Analysis
The attached link is a downloadable copy of the above tables which serve to further show additional planning, work and calculations. KEY
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||