BME103:T930 Group 14
| Home People Lab Write-Up 1 Lab Write-Up 2 Lab Write-Up 3 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
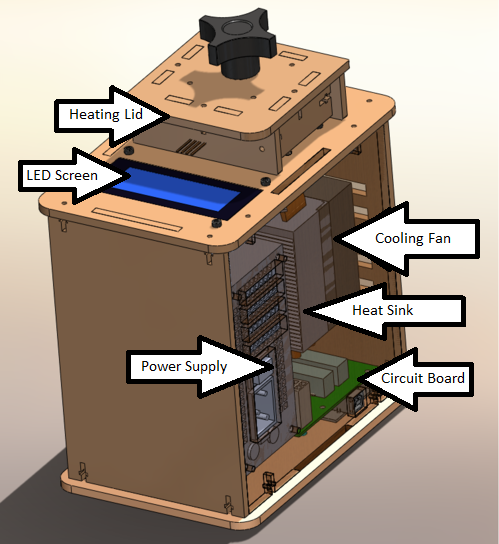
OUR TEAMLAB 1 WRITE-UPInitial Machine TestingThe Original Design Description of Open PCR Device The Open PCR device is thermo-cycler that helps to create strands of DNA. PCR stands for polymerase chain reaction. The Open PCR consists of a heating lid that clamps down on mini test tubes that are held 16-tube PCR main heating block. It also has a cooling fan to help bring down the temperature when it is necessary in the running experiment. The Open PCR alternates between temperatures in order to properly separate deoxyribose nucleic acid (DNA) strands, let primers bind to these strands, and then finish replication of DNA. Then the process repeats for several cycles. The LED screen displays the temperature, the cycle number over how many cycles total, and the ETA time until the test in completed. It heats up to 110 degrees Celsius by default, while its cool temperature sits at 25 degrees Celsius. The Open PCR device must be plugged into the wall (power source) and can be connected to a computer via a USB port. The device is controlled by a simple software on the computer, which allows for the temperatures and cycles to be changed as needed, as well as a start/end test button.
When we unplugged the LED from the Open PCR circuit board, the machine stop displaying information on the LED screen. When we unplugged the white wire that connects the Open PCR circuit board to main heat sink, the machine incorrectly displays the temperature as negative 40 degrees Celsius. Test Run The first test was an experimental run to be sure the machine was running properly. Empty PCR test tubes were placed in the main heating block and the heating lid was shut closed. The Open PCR communicated efficiently with the computer, starting when the "start test" button was clicked. The Open PCR heated and cooled properly, therefore passing guideline markers that the device was running properly. This test was conducted on October 25th, 2012 and the test was successful.
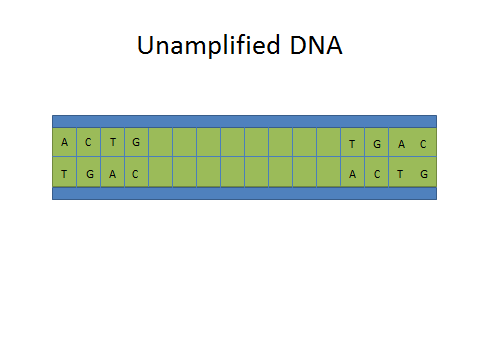
ProtocolsPolymerase Chain Reaction The Polymerase Chain Reaction is a process that relies on thermal cycling allowing the DNA to be broken apart and replicated. This biochemical process works by separating a strand of DNA allowing a primer to target and begin replicating a segment of DNA. This separation, replication, and amplification of a specific DNA strand occurs within the machine during the thermal cycling.
PCR Master Mix Components GoTaq® Colorless Master Mix, 2X Upstream primer, 10μM Downstream primer, 10μM DNA template Nuclease-Free Water Note: GoTaq® DNA Polymerase is supplied in 2X Colorless GoTaq® Reaction Buffer (pH 8.5), 400μM dATP, 400μM dGTP, 400μM dCTP, 400μM dTTP and 3mM MgCl2.
The Samples
Fluorimeter Setup  
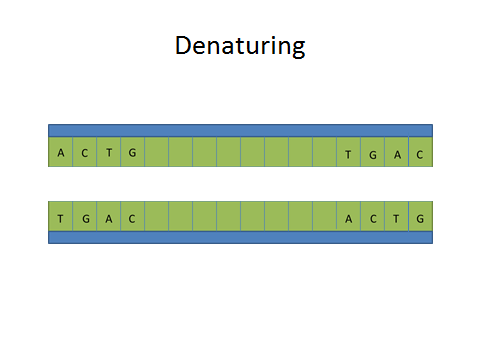
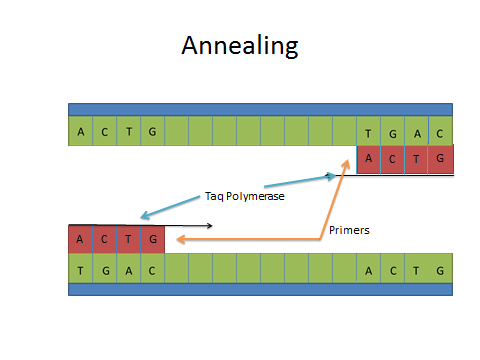
Research and DevelopmentSpecific Cancer Marker Detection - The Underlying Technology To understand how a Polymerase Chain Reaction machine works, first an understanding of how DNA replicates in the human body must be explained. DNA must replicated to form new identical cells so that the organism the cells are supporting can grow and adapt to conditions it faces. The replication process is very complicated and involves a series of enzymes. In a nutshell, DNA begins with a double helix structure. Once unwound, by the enzyme helicase, the base pairs (Adenine, Thymine, Guanine, and Cytosine) that were held together by hydrogen bonds are now exposed and able to bind with free floating dNTPs with the help of DNA Polymerase III after a primer has been laid down. The step of adding a primer is extremely important. Without an initial strand of RNA to allow DNA Polymerase III to bind, replication would not occur. This is extremely important for our experiment involving the PCR machine. We are testing to see if a cancer gene is present in patients, and without the initial DNA primer being able to bind, we can tell if the cancerous genes are present in that person because the DNA would not be amplified. More on that later. When using an Open PCR (Polymerase Chain Reaction) machine for the first time, you must first make sure that you have a computer capable of running the software and download it. After the software is loaded, plug in the Open PCR machine and prepare your DNA samples that were submitted to be tested for the cancer marker. First, you must prepare the samples for amplification by adding the sample DNA, Taq Polymerase (for replication), MgCl2, and the dNTPs. This experiment focused on identifying a cancer associated gene (rs17879961) that was the result of a missense mutation (a change in the sequence ATT to ACT) which would cause the protein transcribed to become threonine instead of isoleucine. That sequence is related to patients getting Breast Cancer and Colorectal Cancer. The DNA primer being used for the detection of this cancer sequence is going to be AAACTCTTACACTGCATACA, which has the missense mutation within it. The way to know if the sample DNA has a positive result for the cancer gene is if amplification occurs. The primer will bind to the DNA if the cancer sequence is present and the strands will replicate exponentially, whereas if the cancer marker is not present, the primer will not bind to the DNA and it will not replicate because Taq Polymerase will not be able to bind. After the samples are prepared for amplification, the PCR program must be set up. To mimic the replication process of DNA in human cells, we manipulate the system by adding Taq Polymerase instead of DNA Polymerase III because the Taq Polymerase is heat resistant, and heat is going to be used to take place of the enzymes that originally initiate and carry out the DNA replication process. The temperatures that need to be programmed into the PCR are 95˚C (separates the DNA strands to prepare for replication), 57˚C (causes the primers to bind to the DNA), and 72˚C (causes Taq Polymerase to be initiated). There should be 30 cycles entered into this program to make sure ample amplification has occurred. After the 30 cycles are complete, the user can take out the sample and freeze them for later use. When the reaction is finished, the samples can be tested to see if the cancerous gene is present. As discussed before, the primer has to bind with a very specific section of the DNA to initiate replication (Adenine can only bind to Thymine and Guanine can only bind to Cytosine). If the DNA has been replicated exponentially and a large quantity of the target DNA now lies within your sample, you know that the specific primer matched with the DNA strand and the cancerous gene is present. If no replication has occurred and a small amount of DNA is still in the sample, however, you can conclude that the DNA is free of the cancer gene because the primer could not match up with the strand. That would make Taq Polymerase unable to bind and add the dNTPs so the DNA could be copied. When looking at the statistical significance of the data, Bayesian Reasoning was applied, which is: [math]\displaystyle{ P(CT) =\frac{P(TC)\, P(c)} {P(TC)\, P(c)+P(TC)\,P(c)}. \, }[/math] . The proportion of patients with positive results is the proportion of A compared to A+C, where the top is A and the bottom portion is A+C. If applied to a group of 10,000 patients, where 1% have cancer, 80% of people with cancer will get a positive result, and 9.6% will also get a positive result, we found that the frequency of this cancer: 80/(80+950) = 7.8%.
Results 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||