BME100 s2014:W Group5 L4
OUR TEAM
LAB 1 WRITE-UP
Machine Troubleshooting: Understanding How OpenPCR Works
Initial Design
http://makezine.com/2010/06/24/things-heat-up-for-openpcr-project/
Manipulating the Connections
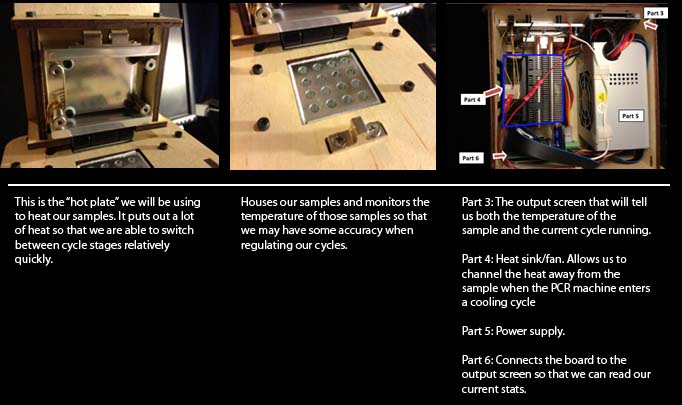
Unplugging part 3 from part 6 removes both the power source and the input to the screen, so it shut off.
Unplugging the white wire cuts the connection between our "thermometer" and the board. If we maintain the
connection between the board and the output screen but unplug the white wire, the display reads the default temperature value.
Part 1::
Heat source:
If this part didn’t work, we would be
unable to heat our samples.
Part 2::
Sample hub:
This part houses the sample and monitors the
temperature. If this part didn’t work, the samples would
not be regulated and all ability to manipulate the
temperature of the sample would be lost.
Part 3::
LED Screen:
When disconnecting part 3 from part 6, the LED screen shut off due to the fact that part 6 is the circuit board.
Part 4::
Heat Sink/Fan:
This part regulates the temperature of the unit
mechanically in response to the digital commands
given by the OpenPCR software.
Part 6::
Circuit Board:
The output screen did not receive a reading for the
temperature when disconnected from part 2. This
should be expected since the sample temperature is read
from part 2.
Test Run
Settings:
HEATED LID: 100°C
INITIAL STEP: 95°C for 2 minutes
NUMBER OF CYCLES: 35
Denature at 95°C for 30 seconds
Anneal at 57°C for 30 seconds
Extend at 72°C for 30 seconds
FINAL STEP: 72°C for 2 minutes
FINAL HOLD: 4°C
The machine that was used in the test run was not able
to pass one cycle. Since it is expected for PCR to
run at least twenty cycles, the machine was
marked as "fail."
Protocols
Thermal Cycler Program
Stage:
1
INITIAL STEP: 95°C for 3 minutes
Denature at 95°C for 30 seconds
2
Anneal at 57°C for 30 seconds
3
Extend at 72°C for 30 seconds
FINAL STEP: 72°C for 3 minutes
FINAL HOLD: 4°C
DNA Sample Set-up
Positive Control (+):
diseased DNA
Negative Control (-):
non-diseased DNA
Patient 1 (ID:73190):
Label: A1, A2, A3.
Patient 2 (ID:43566):
Label: B1, B2, B3.
DNA Sample Set-up Procedure
50 microliters of PCR reaction mix in each of the eight tubes.
50 microliters of DNA/primer mix in each of the eight tubes. The type of DNA varies depending on the tube.
PCR Reaction Mix
- Taq DNA polymerase:
GoTaq
- MgCl2
- dNTP's
DNA Sample/Primer Mix
The DNA samples were of different patients that had both a forward and reverse primer.
Research and Development
Specific Cancer Marker Detection - Understanding the Process
PCR, or polymerase chain reaction, is a synthetic process that catalyzes the replication of a specific strand of DNA.
Function of each component:
Template DNA:
The DNA we start with. It will be the template, if you will, for the replicates.
Primers:
Serves as a starting point for DNA synthesis. Primers attach in the cooling step of PCR. Primers ease the attachment of taq polymerase to the
desired portion of the template DNA. Different primers are used to replicate different portions of a DNA strand.
TAQ Polymerase:
Used in the PCR. Synthesises a new strand of DNA from the template. It works particularly well in high temperatures.
Magnesium Chloride:
Acts as an inert catalyst for the reaction. It aids in the synthesis of many biological compounds like ATP and DNA. The magnesium ion,
specifically, aids in the annealing process as it acts as a bridging complex for electrons to transfer from one nucleotide to its respective
partner.
dNTPs:
Deoxynucleotide triphosphate solutions are utilized in PCR to supply nucleotides(Adenine, Cytosine, Guanine, and Tyrosine) for the synthesis of
new DNA strands. Once these materials run out, the synthesis cannot proceed.
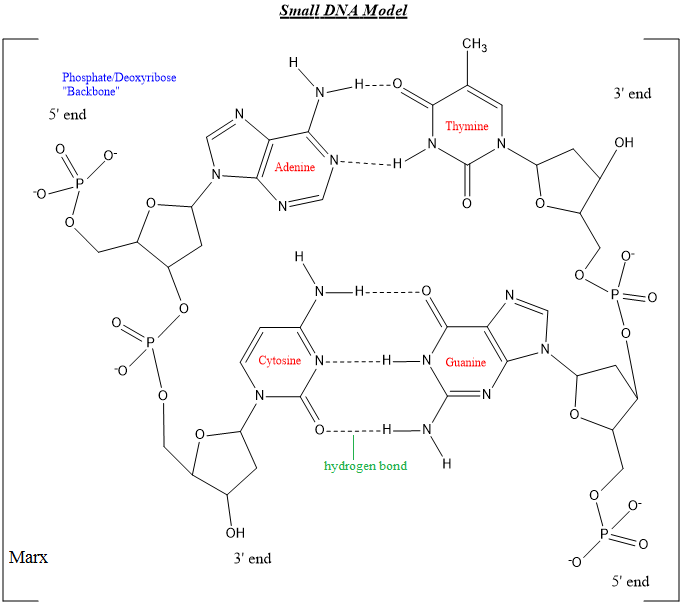
DNA Pairing:
Nucleotide, deoxyribose and phosphates form the complex of DNA shown below. It can be observed that guanine and cytosine have three hydrogen bonds between them whereas the adenine/thymine pair have two. The hydrogen bonding sites are where the denaturing occurs in DNA replication during the first step of PCR.