BME100 f2018:Group7 T1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||
OUR TEAMLAB 4 WRITE-UPProtocolMaterials
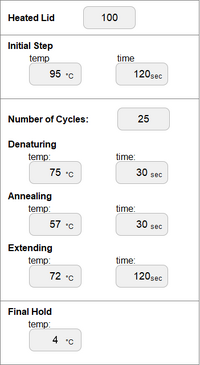
INITIAL STEP: 95°C for 2 minutes NUMBER OF CYCLES: 25
FINAL STEP: 72°C for 2 minutes FINAL HOLD: 4°C PCR INTERFACE EXAMPLE:
Research and DevelopmentPCR - The Underlying Technology
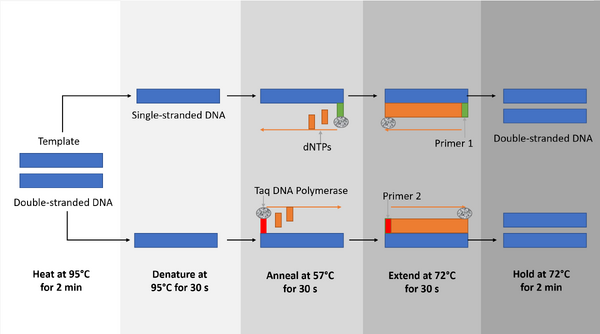
Polymerase Chain Reaction (PCR) is a multistep process that utilizes multiple components, such as primers, template DNA, Taq DNA polymerase, and deoxyribonucleotides (dNTP's); each component has a specific function. Primers are short pieces of DNA that are made in a laboratory, and they attach to areas on the DNA strands at both of the ends of the segment you want to copy, with a low probability of identifying the wrong site. Taq DNA polymerase is an enzyme which preserves a genome during replication and is also a thermophile that can survive extremely high temperatures. Taq DNA polymerase is used in PCR due to the fact that DNA is heated to high temperatures during the PCR process; thus, an enzyme that is capable of withstanding high temperature is required. Deoxyribonucleotides are the four molecules (Adenine, Cytosine, Guanine, Thymine) that make up the DNA structure. The template DNA is the DNA that serves as the basis for replication during the PCR process.
At the initial step, when the DNA is heated to 95°C for 2 minutes, the DNA double helix unwinds, followed by the denaturing process, which creates two single strand DNA molecules. Upon completion of this step, the anneal process at 57°C for 30 seconds allows for single-stranded DNA molecules to attempt to pair up, but the primers in the tube lock their target before the strands can rejoin. During the extending process (at 72°C for 30 seconds), the temperature activates the DNA polymerase, which locates a primer attached to the DNA strand and begins to add the complementary nucleotides to the strand until it reaches the end. During the final process (a 72°C hold for 2 minutes) the two strands of the desired product begin to appear. Through the repetition of this process, the two strands will be multiplied into many. The final hold at 4°C is designed to maintain the PCR product until use.
Nucleotide base pairs anneal to complimentary base pairs through a hydrogen bonding process. The base pairs anneal in a specific manner - Adenine will always pair with Thymine, and Guanine will always pair with Cytosine. Similarly, Thymine will always anneal with Adenine, and Cytosine with Guanine.
Nucleotide base pairs anneal to their complimentary counterparts during the extending process of thermal cycling, as well as the anneal process. The extending process allows for the DNA polymerase to produce the complimentary nucleotides to the strands of DNA, thus completing the sequence.
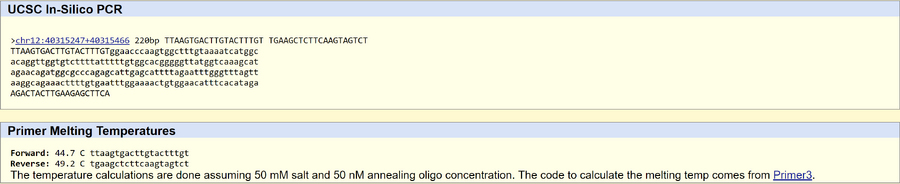
SNP Information & Primer DesignBackground: About the Disease SNP Nucleotides are the building blocks (monomers) for nucleic acids. There are four types: Adenine (A), Thymine (T), Cytosine (C), and Guanine (G). A genetic polymorphism is a variance in the DNA. Thus, a SNP is a difference in a single nucleotide. This specific SNP being examined is found in Homo sapiens. It is located on chromosome 12:40315266. Clinical significance is unknown but according to PubMed, it is linked with familial parkinsonism. LRRK2 stands for leucine rich repeat kinase 2. The functions of LRRK2 include: ATP binding, GTP binding, and GTP-dependent protein kinase activity. An allele is a variant form of a gene located on a specific chromosome and at a specific position. The disease-associated allele contains the codon GAG. The SNP causes a change from Valine (GTC) to Glutamine (GAG). The numerical position of the SNP is 40315266.
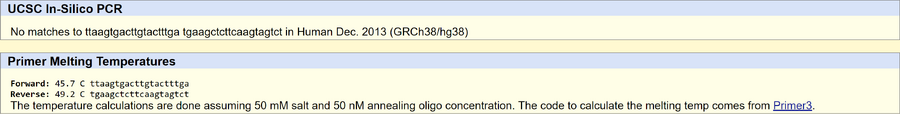
Primer Design and Testing The non-disease forward primer (20 nt) is 5'-TTAAGTGACTTGTACTTTGT-3' The non-disease reverse primer (20 nt) is 5'-TGAAGCTCTTCAAGTAGTCT-3' The disease forward primer (20 nt) is 5'-TTAAGTGACTTGTACTTTGA-3' The disease reverse primer (20 nt) is 5'-TGAAGCTCTTCAAGTAGTCT-3' The UCSC In-Silico PCR tool validated the results. The non-disease sequence was found in the human genome and the disease sequence was not because it is a mutation.
|
||||||||||||||||||||||||||||