BME100 f2018:Group7 T0800 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||
|
OUR TEAM
PCR 2.0 LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System The BME100 class was able to test a sample of 34 patients to find out if the disease-associated SNP was a component of their genes. The results were analyzed, separated and labeled as positive, negative, or inconclusive, with both positive and negative controls to ensure appropriate scientific practice. Also, through collecting similar data as a class, multiple trials were conducted to increase the validity of the result. However, to further prevent error, each patient had two replicates, ensuring that test failure for that particular patient would be negligible due to repeated tests on that same individual. Also, when taking pictures and analyzing through ImageJ, we made sure to hold the camera in place for each of the three pictures per PCR sample, and proceeded to measure the distance between the lens and droplet for each sample. Finally, we outlined each droplet to separate it from the background and enable ImageJ to provide the most accurate results. From the final data after conducting all of the PCR tests, there were only three inconclusive conclusions, with eleven positive and the rest negative. Although there were more inconclusive tests for each patient, the overall conclusion ended up leaving less "blank data" which was easier to interpret and more accurate in comparison to the real results, leading into the Bayes statistics analysis. As far as challenges go, there might have been some discrepancies in our PCR data collection, as we switched off with the micropipettor and ended up with slightly different amounts of solution left when going into the waste bin. This would've had a minimal but not statistically irrelevant impact on our results after performing PCR. However, we think that by eliminating other possible confounding variables and focusing on accurately performing the experiments and tests, our results are consistent enough to be reliable. What Bayes Statistics Imply about This Diagnostic Approach
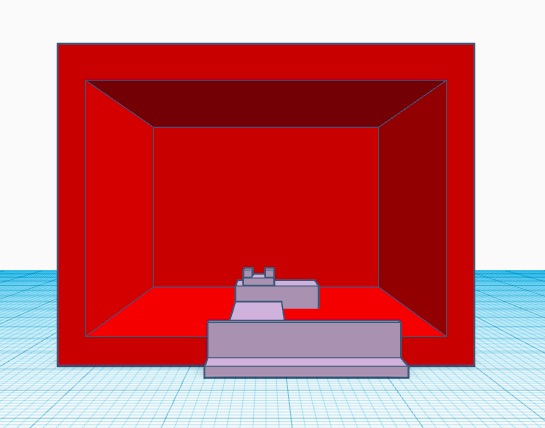
THREE POSSIBLE SOURCES OF ERROR During the wet lab, the process to prepare our patients' DNA data through the PCR could have resulted in a lot of human error. One possible human error could have been when micropipetting the PCR solutions into the tubes to put into the thermal cycling. It was most of the group's first time micropipetting and there could be some user error in not getting the measured amount of solution that supposed to be extracted and when moving the solution to one tube to another could have resulted in loss of the solution in the micropipetting tip. Another source of error could have been with using the ImageJ program to get data points. We had to draw an oval around the droplet to be able to measure it and then move it up to the background to subtract the two. ImageJ was rather very finicky in that the ovals around the droplets could have lend to insufficient data at the end. The last possible error could have been the camera that we used. The camera is an essential factor in how we got our data points as the camera captured the pictures and we ran them through ImageJ. If the camera was too far away or not focused enough that could compromise the accuracy of our photos and evidently the accuracy of our data points. Intro to Computer-Aided Design3D Modeling The software used to make our 3D model was TinkerCAD. Our experience with using this software was a little hard to control or adjust the parts because we didn't have a computer mouse. It made it difficult for us to zoom in and out to see if the parts were connected correctly especially when rotating parts around it was hard to align correctly with the others. While making our 3D model we did some research on TinkerCAD to find the shortcuts to make it easier to construct our model. After watching a couple of videos it became fairly easy and noticed that TinkerCAD is a bit easier to use then Solidworks. The thing that made it easier was having basic shapes such as cubes, spheres, and cylinders prepared whereas Solidworks you have to start from scratch. All we did was drag the shape that we wanted onto the grid and changed its dimensions to correspond to what we wanted our model to look like. To group our parts together TinkerCAD is easier to use for this because all we had to do was just highlight the objects, click group, and everything would be connected. As for Solidworks, you have to click on the faces that you want to be grouped together. Also, TinkerCAD has a variety of colors to chose from to make specific parts stand out from others. TinkerCAD is easy to use and to navigate. Our Design Our design was altering the original design of the fluorimeter set up. We chose to alter this design, because we struggled to get the placement right on the set up during the lab. We changed a couple things, instead of the box being open at the bottom we included a platform for the actual fluorimeter to sit on and stay stationary. So when we lift up the box we know exactly where to place the box back over. Then we extended a second platform from the fluorimeter to a stand for the camera. The camera placement was essential in getting a clear picture and the replication was important. Instead of having the camera stand separate from the fluorimeter, we have attached the two so we can replicated it precisely. Another alteration we have made that is not displayed is the use of a better quality camera. The iphone is hard to focus on and change settings that you could on a normal camera. You would have to go into settings and find what camera settings were best for this, so instead we would use a camera that had readily accessible camera settings. Our struggles in the lab was the purpose behind all our alterations to the fluorimeter. With these alterations, we can capture better quality pictures and better results for the lab.
Feature 1: ConsumablesOur PCR Kit includes PCR tubes, primer solution, SYBR Green solution, Buffer, Hydrophilic glass slides, 100 microliter micropipette tips and ICY Bioimages photo analysis software.
Feature 2: Hardware - PCR Machine & FluorimeterWe will use the Open PCR machine in order to obtain our desired DNA strands. However we have excluded the fluorimeter set up as it was very difficult to use. We have designed a new fluorimeter system in which all components are attached and do not require set up. We will still use the same glass slide holder and box however they will not be free moving. We have used a different camera stand as our iPhones were difficult to focus and did not produce the quality image we desired. We designed a camera stand that was compatible with higher power camera capable of manually focusing. We decided to modify our fluorimeter as we did have the most issues with this hardware due to all the individual components that were easy to move and required readjustment after every trial.
| |||||