BME100 f2018:Group7 T0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
2, and dNTP’s
have the same forward primer and reverse primer
Research and DevelopmentPCR - The Underlying Technology Components of PCR Reaction PCR, or polymerase chain reaction, is a method of copying DNA molecules. The goal of the PCR reaction is to replicate only a portion of the genome of interest. There are different components that complete separate functions in the PCR reaction: template DNA, Primer, Taq Polymerase, Deoxyribonucleotides (dNTP’s). A template DNA is the DNA that is going to be replicated. In order to bind to the template DNA at desired points so that the polymerase can replicate the desired segment and prevent the template DNA from rejoining at cooling temperatures, a primer is needed in the PCR reaction. After the primer, the taq polymerase is used to replicate the desired segment of template DNA. Deoxyribonucleotides is the monomer, or single unit, of DNA and it is able to take in the polymerase in order to be organized in a replica of the template DNA. Steps of Thermal Cycling In the initial step, the DNA template begins to separate into two single stranded DNA at 95°C. After 2 minutes, the thermal cycler cools down to 57°C for 30 seconds in order for the primer to bind onto a specified point and with the primers in place, the single strand DNA are not able to bind together as they would naturally without the primers in place. Then the thermal cycler rises to 72°C and within 30 seconds the Taq DNA polymerase locates the primers and is activated then begins to replicate the desired segments of DNA. Now the first cycle is complete. Cycle one is repeated for cycle two and for cycle three you start to obtain your desired product. Two strands of DNA with primer one on one end and primer two on the opposite end. Then the thermal cycler is cooled down to 4°C to prevent the primers and taq DNA polymerase from being activated and replicating any nonspecific sequences. The final step is to increase the thermal cycler to 72°C for 2 minutes; after cycle three you have isolated the desired segments of DNA which you wish to replicate therefore you can hold the temperature at 72 degrees celsius to allow taq DNA polymerase to operate without interruption and replicate the desired segments of DNA without any extra nucleotides. Four Types of Nucleotides Base‐pairing, driven by hydrogen bonding, allows base pairs to stick together. The nucleotide Adenine binds to thymine and vice versa, while cytosine binds with guanine and vice versa. In the PCR reaction, the two steps that occur in 72°C toward the end of the thermal cycling base-pairing is usually occurring between and within these steps.
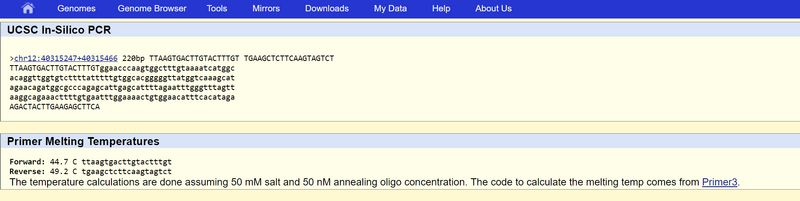
SNP Information & Primer DesignBackground: About the Disease SNP An SNP (Single Nucleotide Polymorphism) is a genetic variation in people, each SNP represents the difference in a single DNA block called the nucleotide. The disease associated with the SNP is the Leucine-rich repeat kinase 2 gene (LRRK2), which is located in the chromosome specifically the 12:40315266. The Leucine-rich repeat kinase 2 gene encodes a protein with an ankryin repeat region, a kinase domain, and a variety of other domains. The protein is located in the cytoplasm and is related with the mitochondrial outer membrane. The condition that correlates to the SNP is Parkinson's Disease and the numerical position of the SNP is 40315266. The disease-associated allele contains a codon of GAG.
The forward and reverse primer both match because of every PCR needs two primers to magnify the DNA. The non-disease forward and reverse primer both matched because the chromosome was 220bp from the disease SNP. The non-disease forward primer 5'-TTAAGTGACTTGTACTTTGT-3'. There was a no match when the forward primer was changed to a pair of disease SNP primer. It was changed because the disease SNP primer had to be the same as the SNP nucleotide (disease). This proves that our primer sequences were correct. |
||||||||||||||||||||||||||||||||