BME100 f2018:Group3 T1030 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||
OUR COMPANYOUR TEAM
LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System In the BME 100 lab, there are 17 teams of 4-6 students each who were assigned to test patients for a disease-associated SNP. This was done by each team being given two patient samples with different identification numbers indicating that their contents were not the same. With these samples, each team conducted PCR with three replicates (samples) per patient as well as the positive and negative controls. Following PCR, these results were then analyzed using a fluorimeter to take pictures of the illuminance. Out of these pictures, we used three of them for ImageJ to calculate and help guide us to determine if our patient samples were positive or negative for the SNPs. After looking at the data, we found that both of our patient samples were negative. Overall, the lab was fairly simple and did not have many challenges. There was not much room for error and it didn’t feel as though our group could have encountered any situations that would have affected the data. There is always room for error but if any was made, it was so minute that the effect would invisible. What Bayes Statistics Imply about This Diagnostic Approach The results from calculation 1 is close to 1.00 (100%). This means that the probability of getting a final positive test conclusion given a positive PCR reaction is close to 1.00. Calculation 2 indicates that the probability of getting a final negative test conclusion given a negative PCR reaction is close to 1.00. The results from calculation 3 is close to .50 (50%). This refers to the accuracy of the system to predict if the patient develop the disease but only to 50% accuracy. Calculation 4 refers to the the accuracy of the system to predict if the disease develop was more reliable, 100% accuracy. Several errors includes the inaccuracy in measuring the samples and solutions throughout the lab. Another is the pictures taken might be low quality and the slight shift in the pictures, this would cause ImageJ to display the incorrect values. Last possible error was that the SYBR was exposed to light every time it is used, this would make the later tests be more unreliable.
Intro to Computer-Aided Design3D Modeling Our Design
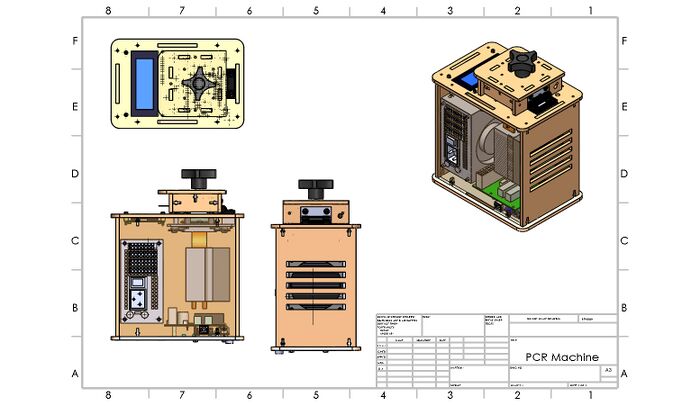
During the progress of developing the PCR machine there are many encounters that can lead to miss calculations. Our top priority was to develop a machine that would allow anyone to analyses DNA to establish its principles. To keep things as simple as possible, the redone PCR machine is an all in one machine that is able to go through the PRC reaction, adding in the SYBR Green 1 Solution, buffer and water. Again able to heat up for the appropriate temperatures for the correct reactions. The final use of the redone PCR machine would be to take pictures of the glow of the substances and give the appropriate readings from the results.
Feature 1: ConsumablesPCR reaction on mix, 8 tubes, 50 μL each: Mix contains Taq DNA polymerase DNA/ primer mix, 8 tubes, 50 μL each: Each mix contains a different template DNA. Each with the same forward and reverse primer A strip of empty specialized PCR tubes, only to be used in cooperation with our brand of PCR machine Disposable pipette tips 1 Micropipette OpenPCR machine 2 tubes of 1000 μL of SYBR Green 1 Solution. 1 tube of 500 μL of Buffer. 1 tube of 1000 μL of water at pH=8 5 tubes labeled 0.25, 0.5, 1, 2, and 5. These contain Calf Thymus
Feature 2: Hardware - PCR Machine & FluorimeterThe redesign of the PCR machine is meant to be an all in one take on the current design of the current model. The tubes used for the redesigned machine will be specially made to fit this machine only. The redesign of the PCR machine will be able to go through the PCR reaction, add in the predetermined amount of SYBR green and allow for the correct amount of time. The SYBR green is to be added in after PCR reactions takes place like normal. After all the solution's have been properly combined the redone machine will be able to use the fluorimeter. On the outside of the PCR machine a USB ports will be present to be able to transfer the given photos and data. This change to the hardware is meant to be an easy and simplified version of all the other current device's as this removes the need of having to swap out all the solutions from one place to another, and can assure a significant reduction in margin of error.
| |||||