BME100 f2018:Group3 T0800 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||
OUR COMPANY
LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis SystemThe BME 100 class tested patients for the disease-associated SNP by using a polymerase chain reaction (PCR). There were a total of fifteen teams of five to six students. Each team diagnosed two patients, therefore the entire class diagnosed a total of thirty patients. There were many preventive steps taken to minimize the possibility for error. First off, the baseline positive control and negative control each had three pictures taken to be run through ImageJ. The average was taken of the three values. As for the patient samples three samples were taken from each patient for a total of six samples. Each sample had three pictures taken and run through ImageJ. The average of those three values was also taken for that sample. In total 24 images were run through ImageJ. The ImageJ calibrations were controlled as was the number of drop images that were used for the ImageJ calculations. In terms of running DNA tests, the class completed 90 total individual PCRs (not including the controls) where 30 of the total 90 were positive, 44 of the total 90 were negative, and 16 of the total 90 were inconclusive. The class had come to a total of 30 completed conclusions where 11 of those total 30 completed conclusions were positive, 16 of the total 30 completed conclusions were negative, and 3 of the total 30 completed conclusions were inconclusive. In total, it was determined that there were 25 positive PCRs given that there was a positive conclusion and 29 negative PCRs given that there was a negative conclusion. In terms of diagnosis, there was a total of 11 total positive conclusions and 16 total negative conclusions. There was total of 10 "yes" diagnoses and 20 "no" diagnoses. There was a total of 4 positive conclusions given there was a "yes" diagnosis and a total of 11 negative conclusions given there was a "no" diagnosis. What Bayes Statistics Imply about This Diagnostic ApproachCalculation 1 describes the sensitivity (the probability that a person with the disease being tested with test positive) of the system regarding the ability to detect the disease SNP. The results for Calculation 1 implies that there is a high reliability of the individual PCR replicates for concluding that a person had the disease SNP. The Bayes value found for Calculation 1 was between 0.9 and 1.00 (90-100%). Calculation 2 describes the specificity (the probability that a person who does not have the disease being tested for will test negative) of the system regarding the ability to detect the disease SNP. The results for Calculation 2 implies that there is a high reliability of the individual PCR replicates for concluding that a person does not have the disease SNP. The Bayes value found for Calculation 2 was between 0.95 and 1.00 (95-100%). Calculation 3 describes the sensitivity (the probability that a person with the disease being tested with test positive) of the system regarding the ability to predict the disease. The results for Calculation 3 implies that there is a low reliability of PCR for predicting the development of the disease (diagnosis). The Bayes value found for Calculation 3 was less than 0.5 (50%). Calculation 4 describes the specificity (the probability that a person who does not have the disease being tested for will test negative) of the system regarding the ability to predict the disease. The results for Calculation 4 implies that there is a mid-ranged reliability of PCR for predicting the development of the disease (diagnosis). The Bayes value found for Calculation 3 was between 0.5 and 0.75 (50-75%). The first possible source of error that could have occurred during the PCR and detection steps that could have affected the Bayes values in a negative way could be human error with the micropipetting. The way that the liquids were micropipetted could have been different every time which could have affected the results. The second possible source of error that could have occurred during the PCR and detection steps that could have affected the Bayes values in a negative way could be human error with the ImageJ. The experimenters took care in making sure the ImageJ calculations were as accurate as possible, however there could have been error when running the program. The third possible source of error that could have occurred during the PCR and detection steps that could have affected the Bayes values in a negative way could be the PCR machine itself. The experimenters have little control over the PCR machine itself and left it to run without monitoring it. There is a possibility that someone could have tampered with it. Intro to Computer-Aided Design3D ModelingFor our PCR reaction we found that the main issue we encountered was that the phone stand kept falling over. In order to redesign the phone stand we used SOLIDWORKS in IPS format. The process to redesign the stand was not too difficult because many of our team members already had experience with this software so we were able to do different parts of the stand and then one of our team members assembled the whole thing. In order to effectively make an assembly we had to make sure that the measurements were the same for all the parts and that the extruding and cutting actually matched. Although a phone stand sounds pretty simple the design was quite complex because of the multiple parts that went into it. There had to be a container to where the phone would be placed, slides that would adjust the holder based on the phone size and the base, which was composed of two different parts, one for each side. Our Design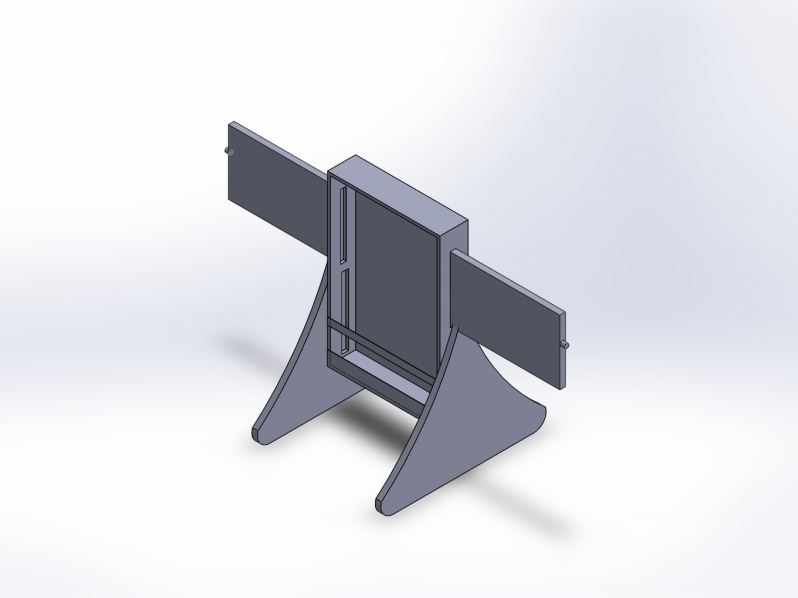
Feature 1: ConsumablesThe consumables kit will include all the liquid reagents. The liquid reagents included are: PCR mix, primer solution, SYBR Green solution and buffer. The kit also includes the plastic tubes, glass sides, and pipettors and pipette tips. The new addition to the kit will be a new phone stand. The phone stand will not be as fragile as the previous stand from the last lab since it will now be able to stand still without it falling over. Feature 2: Hardware - PCR Machine & FluorimeterThe same Open PCR machine and fluorimeter that was used previously will be included in the new system. The experimenters did not find any faults with the Open PCR machine and the fluorimeter when working with it previously, therefore did not see a need to fix it. They will be incorporated as they were before as the only thing that is being improved upon is the phone stand that will hold the phone to take pictures when using the fluorimeter. Image of Feature 1 and Feature 2
| |||||||





