BME100 f2018:Group14 T0800 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||
OUR COMPANY
Planck PCR LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System In BME 100, the students were split into 17 groups with five to six students in each. Each of the teams was assigned two patients, and the task at hand was testing for a disease-associated (Parkinson's Disease) SNP in DNA using an OpenPCR machine. In this machine, the DNA samples underwent a polymerase chain reaction, meaning the DNA was replicated and specific parts were amplified. To prevent error and confusion, each PCR tube was given a unique label. Because this was an experiment, a positive and negative control were given to compare other results against. To allow for a larger sample size and ensure accuracy, each team was given three samples of DNA from each patient, and three images from each sample were taken and analyzed using ImageJ. The mean was then taken of the three images for each sample before computing results. In total, the class performed 90 imaging analyses and of the 90, 30 produced positive results, 44 produced negative results, and 16 were inconclusive. Of the 30 patients tested, the class found 11 to be positive, 16 to be negative, and 3 to be inconclusive. This differs from the actual results, as 10 were positive and 20 were negative. Overall, the PCR results were about 50 percent accurate when compared to the doctor's diagnoses.
During the Bayes calculations 1 and 2 the results for each came close to 1.00 (100 %), with calculation 2 being slightly more accurate. This does indicate that the PCR results show that high probability of a positive test, given a positive PCR reaction as well as a negative test given given a negative diagnostic signal is high. This analysis indicates that sensitivity to both detect a disease SNP and predict this disease is also probable. However, the real-world reliability of the experiment is lowered considerably due to the limited amount of patient testing that has been conducted. Nevertheless, the results would become more clear as more patients are tested to ensure statistically accurate results. Within calculation 3 the results were less than 50% for those who tested positive for SNP, which indicates that the results were rather inaccurate when isolating a targeted disease/DNA identifier. However, in calculation 4 the results were slightly less than 1.00 (100%) for those who tested negative, which indicates that the device is accurate to measure a targeted disease/DNA identifier of those who are negative. Within the realm of human or device errors, there could be issues in setting up the fluorimeter, taking consistent images through the fluorimeter, or ensuring that the PCR machine is operating properly. The fluorimeter was slightly difficult to set up, as the materials involved in the process are so simplistic that it is easy to make a mistake and not initially recognize an issue with the system. Furthermore, it was hard to take precise photos with the set up, as there is a huge amount of room for error in measuring the distance, angle, and consistency that the images are being captured with. We identified this as our primary source of error. However, it is also possible to have problems not involved with the fluorimeter at all, but rather the PCR machine. The machine also presents itself as somewhat simplistic, so it is hard to look at the system and see if it is set up properly, or if something could be broken or missing. Intro to Computer-Aided Design3D Modeling
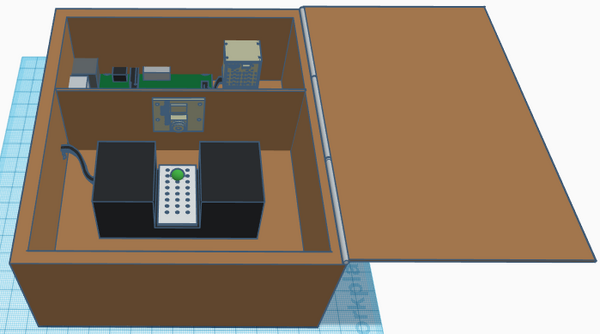
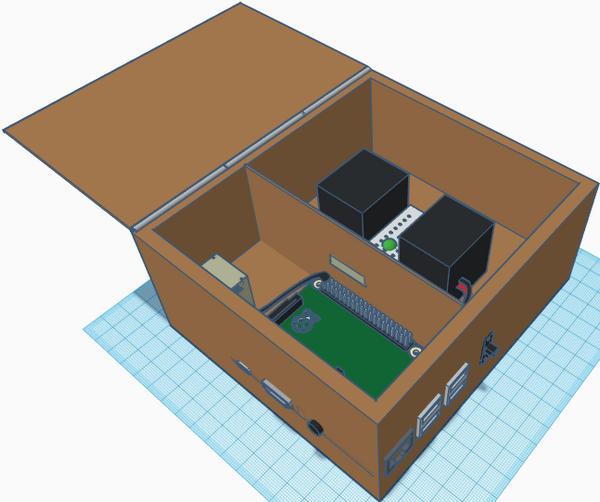
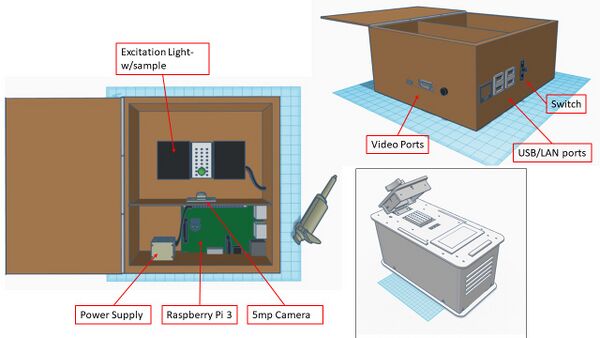
Our overall goal was to vastly streamline this process via multiple design efficiencies, while simultaneously incurring as little cost as possible. Our product consists of a lightweight polycarbonate box containing a Raspberry Pi 5mp camera (mounted on an internal divider to ensure separation of electrical components and PCR reaction liquids), a Raspberry Pi 3, model B+, for control and interface with ImageJ and a hard-point for fixation of the provided excitation light. The entire system can be powered either by the provided power supply, or via any of the USB connections.
Feature 1: ConsumablesAs our group focused more on the fluorimeter-lightbox rather than PCR machine, the group did identify improvements with the consumables issued for the lab. The primary improvement consists of packaging all consumables in kits to be issued to each group. The kits would contain the necessary number of plastic tubes, tube racks, glass slides, gloves, reaction components for the given experiment and a pipette. Tubes would be sized depending on volume requirements and mulitchannel pipettes would be issued if the experiment called for multiple samples consisting identical reaction components. This would greatly streamline the issuing of lab materials and the overall experiment process. Feature 2: Hardware - PCR Machine & FluorimeterAs stated above, our product was designed to be efficient and inexpensive. Every component is either commercial-off-the shelf or easily manufactured with low-cost materials. The camera is fixed, eliminating the need for readjustment, and manual image retrieval is eliminated as all images are saved directly to the lab computer via USB. Real-time visualization, focusing and image taking of the illuminated PCR drops is achieved by system video (HDMI) and comm-port connections to the lab computer. As the system is interfaced through the lab computer (via the Raspberry Pi components), the product was able to be equipment with a light-proof lid which can remain closed during individual sample imaging, thereby effectively eliminating background light contamination. The OpenPCR reaction machine and associated process used previously remains unchanged (despite some design deficiencies) as the team determined that the greatest efficiencies were obtained through re-design of the the fluorimeter-excitation light components.
| ||||||