BME100 f2018:Group14 T0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials Materials for the lab are as follows:
Heated lid at 100°C Initial step is to heat to 95°C for 2 minutes 25 cycles will be run as follow: Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and Extend at 72°C for 30 seconds Final Step is to set at 72°C for 2 minutes Final Hold is at 4°C
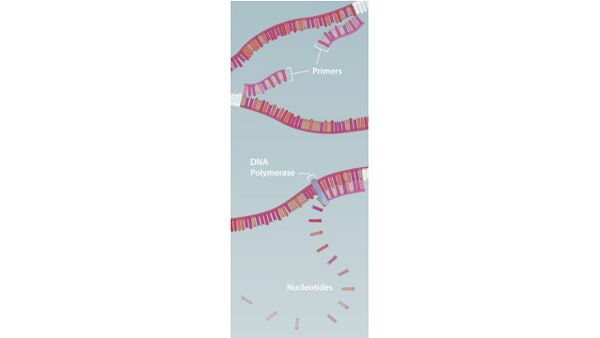
Research and DevelopmentPCR - The Underlying Technology Component Functions in a PCR Reaction In a PCR reaction, there are several components needed, to include a template DNA, DNA primers, Taq polymerase, and, Deoxyribonucleotides. Template DNA is used for the portion of the DNA that is desired to be replicated. Short pieces of DNA, two primer matches to the segment of DNA which we want to copy. One primer will attach to the top of the strand at one end of the DNA segment. Taq polymerase will attach to a DNA template and adds DNA nucleotides to extend the second strand. Deoxyribonucleotides (dNTP's) work to synthesize the specific stretch of double-stranded DNA (A's (adenine) anneals to T's(thymine) and C's (cytosine) anneals to G's (guanine)). Thermal Cycling Effects on PCR Reaction Components DNA goes through certain change during the process of thermal cycling. Initially the sample is heated at 95°C for two minutes. The DNA it gets denatured in 30 seconds at 95°C and the double bonded template DNA is separated into two single stranded DNA. Annealing occurs at 57°C for thirty seconds. During this process the DNA primers attach to each separate template DNA stand. Finally, the temperature increases to 72 °C for two minutes and in 30 seconds DNA polymerase attaches to the primers and DNA nucleotides, creating new strand of DNA. The temperature of the final step is hold for additional two minutes to make sure that bond between the new strand of DNA becomes stable and cools at 4°C. Four Types of Nucleotides Four types of nucleotides make up DNA: A, T, C, and G. The pairing of these bases is fueled by hydrogen bonding. Adenine(A) bonds with thymine(T), while cytosine(C) bonds with guanine(G). Base-pairing Thermal Cycling Steps Base-pairing occurs during the Annealing and Extension steps. After the DNA has denatured into single strands, the system is cooled down to 57°C in the PCR machine and the target SNP is bound with short DNA primers to create new single-stranded DNA. The Temperature rises slightly higher to 72°C so that the thermal cycling stage of extension occurs. The Taq polymers binds to the end of the primer site and extend the primers to form new nucleotide strands of the desired target SNP of the DNA. (see PCR mechanisms image below- source: https://learn.genetics.utah.edu/content/labs/pcr/).
SNP Information & Primer Design
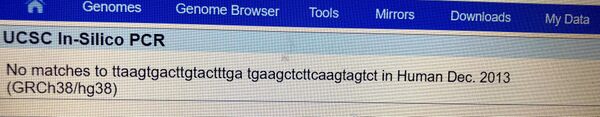
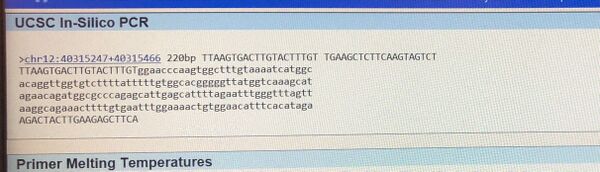
The nucleotides adenine, guanine, thymine and cytosine are the basic structural components of DNA. A polymorphism, the presence of a genetic variation within a population, can result in disease. In Homo Sapiens, the disease single nucleotide polymorphism (SNP) rs721710 is associated with Parkinson's. Specifically, this variation is located on chromosome 12 at position 40315266. Parkinson's is a debilitating neurological disease that can result in muscle rigidity, bradykinesia and tremors. Unfortunately, the exact clinical significance of this SNP is unknown at this time. However, current research is focused on discovering a link between this variation and Parkinson's. Specifically, polymorphisms in the gene LRR2K (leucine-rich repeat kinase 2) have been associated with the disease. This gene is responsible for several functions, including ATP binding, GTP binding and GTP-dependent protein kinase activity. The disease associated-allele (a gene variant) occurs when thymine in the non-disease allele, GTG, is swapped with adenine, resulting in the codon GAG. This polymorphism is located at position 40315266. Primer Design and Testing Designing disease primers for PCR began with identification of the non-disease forward and reverse primers. The forward primer (5’ TTAAGTGACTTGTACTTTGT 3') and the reverse primer (5’‐TGAAGCTCTTCAAGTAGTCT 3’) are both 20 nucleotides long. The forward primer ends at position 40315266, with the beginning 200 nucleotides to the right at position 40315466. To design the forward and revers primers, the last base of the non-disease forward primer was changed to reflect the disease SNP, while the remaining bases were unchanged. The resultant forward primer was 5’‐ TTAAGTGACTTGTACTTTGA 3’ and the reverse primer was 5’‐ TTAAGTGACTTGTACTTTGA 3’ The disease primers were tested against a human genome utilizing the UCSC In-Silico PCR website. The non-disease primers were searched for pertinent matches with the search settings as follows: Genome set to human, Assembly set to Dec.2013 (GRCh38/hg38), Target set to genome assembly, Max product to size to 4000, Min perfect match to 20, Min good match to 20 and Flip reverse primer was unchecked. The non-disease sequence was validated with a 220bp sequence received. The disease sequence was searched in the same manner with a result of "no matches", thus validating that the primer was disease associated as no matches in the human genome sequence were returned (see images below).
|
|||||||||||||||||||||||||||||||||