BME100 f2018:Group12 T0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
Group 12
LAB 4 WRITE-UPProtocolMaterials
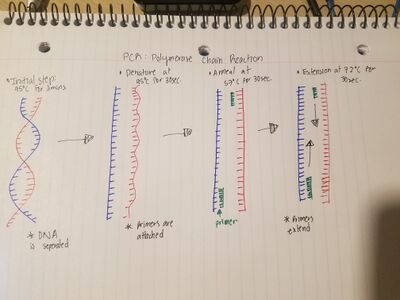
Research and DevelopmentPCR - The Underlying Technology A small sample of DNA can be replicated billions of times in a matter of hours using PCR. To start, the DNA is placed into a PCR tube. The primers, deoxyribonucleotides, and taq polymerase are then added to the PCR tube. The primers will act as boundaries, attaching to either end of the segment of DNA to be replicated. The deoxyribonucleotides are the building blocks for the new DNA being built (A's and T's attach and C's and G's attach), and the taq polymerase will "read" the DNA from primer to primer, and attach the appropriate nucleotides. The PCR process occurs in a thermocycler, which starts by heating the solution to 95°C to separate the double helix. The thermocycler then cools to 57°C, at which point the primers attach to the appropriate section of DNA. Then at 72°C, the taq polymerase activates and attaches to the first primer. It then begins to replicate the DNA strands. Steps of Thermal Cycling DNA goes through many changes throughout the process of thermal cycling. Initially the DNA is heated to 95 degrees C for a total of two minutes almost reaching boiling point. Following for 30 seconds at 95 degrees C denaturing occurs, causing the double bonded DNA helix to break apart into two separate single-stranded DNA molecules. Next, annealing occurs for 30 seconds at 57 degrees C causing the two primers attached to the strand to recombine. The DNA is now extended at 72 degrees C for 30 seconds where DNA polymerase attaches to the primers and DNA stands, adding complimentary nucleotides to the molecule. Concluding the cycle, for two minutes at 72 degrees C The bond between the two strands of DNA becomes stable and cools at 4 degrees C. Base Pairing Base pairing is the classification of DNA, which is made up of four nucleotides A,T,C, and G. Where A's (Adenines) and T's (Thymine) attach and C's (Cytosine) and G's (Guanine) are attached, and occur when the primers attach to the sample DNA, and when the taq polymerase replicates the DNA. Base pairing occurs in both the annealing and extending steps of the PCR cycle.
SNP Information & Primer DesignBackground: About the Disease SNP Single Nucleotide Polymorphisms (SNPs) are the most common type of genetic variation found in human beings. Every SNP is a variation in a single DNA building block also known as a nucleotide. SNPs occur normally in human genes, and in most cases cause no health or developmental issues. Yet they can be used by scientists as biological markers, helping to locate genes associated with disease. Researchers have also found that SNPs can be utilized to predict a patients reaction to a specific drug, their susceptibility to environmental factors, and risk of developing diseases in the future. They can also be used to track the inheritance of certain traits throughout a family tree. SNPs are also being studied further with more serious diseases such as cancer, diabetes, and heart disease to possibly find a cure. Primer Design and Testing
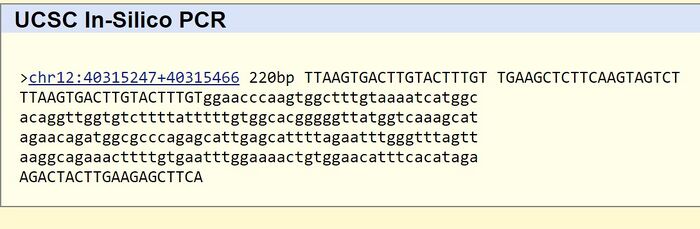
Source: UCSC In-Silico PCR, http://genome.ucsc.edu/cgi-bin/hgPcr?command=start While designing the primer, it was important to search for the forward primer, then the reverse primer. The forward primer was found at SNP 40315266. The reverse primer was found 200 base pairs after the forward primer, at SNP 40315466. Each primer is a total of 20 base pairs. To achieve a mutation, the last base pair mutates into a different base pair. The reverse primer serves as a reference point during the PCR process. The diseased primer returned no results, as a healthy individual is not expected to have the base pair sequence. |
|||||||||||||||||||||||||||||||||