BME100 f2018:Group10 T1030 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||
OUR COMPANY
MAGNE-CLAMP INC. LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System In order for the BME100 class to test patients for the disease-associated SNP that results in Parkinson's, the class divided into 17 groups with about 5 to 6 people per group. Each group diagnosed 2 patients to diagnose a total of 34 patients. In order to prevent error, each patient had three replicates that were tested. We also utilized controls to make sure our results were in range. We did this through the use of a positive and negative controls to compare our final results against. For imagej, we used calibration controls of calf thymus DNA concentrations (5, 2, 1, 0.5, 0.25, 0) so that the quantitative numbers we were finding would be validated. When capturing images, for each calibration as well as each patient replicate, each group took three picture. The results from these three pictures were then averaged and used for our final calculations. In the end, all of the group's final calculations were compiled into a single excel sheet. The amount of data that resulted from the class went down to 32 patients as one group was not able to submit their calculations. In addition, there were 7 patient replicates that were found to be inconclusive and one PCR conclusion that was deemed to be inconclusive. Overall, the class found 14 of the final PCR results to be positive and 17 to be negative. What Bayes Statistics Imply about This Diagnostic Approach
For Calculation 1,our results for the probability of getting a positive test conclusion was a little less than 50% and the probability of getting a positive PCR result was also a little less than 50%. When calculating the probability of a positive PCR given a positive conclusion it was close to 100%. Then, the probability of a positive conclusion given a positive PCR reaction was even closer to 100%. Overall, those who receive a positive PCR reaction result, 95% of them will actually have the SNP disease. Now about 5% will have a false positive. For Calculation 2, our results for the probability of negative conclusion and the probability of a negative PCR reaction were both a little over 50%. Then, the probability of a negative PCR reaction given a negative conclusion was close to 100%. The final probability reading of a negative conclusion given a negative PCR reaction was really close to 100%. Overall, those who receive a negative PCR reaction result, 98% of them will not have the SNP disease. So about 2% have a false negative.
For Calculation 3, our results for the probability of developing the disease is pretty below 50% and the probability of a positive test conclusion is a little less than 50%. Then, the probability of a positive test conclusion given that the patient has the disease is again less than 50%. The probability of developing the disease given the positive test conclusion is really less than 50%. For Calculation 4, the results for the probability of not developing the disease is over 50% and the probability of an negative test conclusion is a little over 50%. Then, the probability of a negative test conclusion given that the patient has developed the disease is over 50%. Finally, the probability of not developing the disease given a negative test conclusion is really close to 1.
Intro to Computer-Aided Design3D Modeling In order to CAD our design, we were given two options on a program to use: Tinker CAD or Solidworks. Although Solidworks is a bit more complicated and difficult to use, this was the program that our group ended up using. Two members of our group, Lauren and Talia, have learned how to use this program in their BME 182 class. Due to this, they felt fairly comfortable using Solidworks. There were a few points in the design process in which we struggled to create what we exactly wanted to see. We also had a little bit of difficulty with the sizes of our individual parts when we were assembling the device; however, we were able to work through our difficulties and complete the designs for the two parts of our product. Our Design
For our glass slide that we re-designed, we wanted to make it easier for scientists or professionals working with PCR mixtures to be able to get the drop of SYBR Green Solution and the other solution onto the slide in the right position. When we were performing this lab experiment, we ran into troubles with the one droplet moving as well as rolling around on the slide. This made it harder for us to properly set the slide with the drop on the light box with the blue LED light shining on the droplet. So, our new take on the glass slide, was to make a dip in it to allow the drop to fall right in and not go anywhere else. It's position would therefore be fixed and would not cause any problems with recording the data and taking a decent picture. This is different from the original OpenPCR design because the glass slide that we used had a flat surface which allowed the droplet to roll right off or not be in the first 2 rows of where it needed to be to take the picture. All we decided to do, was design a concave up dip that would hold our droplet of PCR mix in place, so that we didn't lose any solution from the droplet by it rolling off the slide.
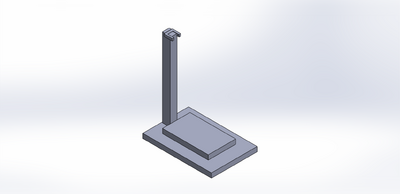
Our main improvement to the whole design and procedure is our PCR Pipette Stand. As we carried out the lab, we took into consideration the fact that you had to be so steady when pipetting the drop on the glass slide. However, it was hard to keep the hand from shaking and therefore placing the drop became difficult. It ended up being in a place different from what it was supposed to be. So our pipette stand was designed to be tall enough for the micropipette to magnetically clamp to the stand and hold it into place. Then, the individual would be able to pipette without shaking or moving the position of the drop. The stand would basically slide right next to light box and the micropipette can then clamp to the magnets.There is a "field-goal-looking" clamp at the top of the stand which would have magnets on the ends that would attract to the magnets on the micropipette. This is different from the original Open PCR design because there is a pipette stand that can help someone that may have shaky hands pipette without any difficulties.
Feature 1: ConsumablesWhen an item is *very important* it means that the item is needed in order to use the device in the kit. Hence, important consumables are components that can be used with our product that only have a one time use. The consumables that would be included in our kit would be glass slides that we redesigned that can be used with our device. We would also include liquid reagents such as PCR mix, SYBR Green solution, and primer solution. We will not include a micropipette, test tube racks, or micropipette tips. The items that we will not be including are fairly standard, so we can assume that the consumer may already have these components or they can easily find them.
Feature 2: Hardware - PCR Machine & FluorimeterOur redesigned components main focus are to be able to use a micropipette with zero errors in the PCR test. The Open PCR machine will still be used to heat/cool the proteins in the DNA. The fluorimeter will still be used to help capture a clear fluorescent color to the small drops; it will still be placed on the slide and shown on by a fluorescent light. Hence, due to out plan to redesign and help with the process of dealing with consumables, we are keeping both the PCR machine and fluorimeter the same. The aim of our product is to ensure accuracy of using a micropipette by eliminating the factor of unsteady hands. With both the micropipette holder and the enhanced slide, the image of the drop should be easier to take. Although our group did not choose to redesign the PCR or the Fluorimeter, we still could find many things that we could change on these machines. Our redesigned components most closely relates to the Fluorimeter process. One thing we realized that was difficult with using the Fluorimeter is that it was difficult to move the light box with slide without shaking the drop on the slide. If we were to redesign the Fluorimeter, we would make it so that the slide and the light box could be moved more efficiently.
| ||||||