BME100 f2017:Group16 W1030 L4
LAB 4 WRITE-UP
OUR TEAM
 |
 |
 |
 |
 |
Protocol
Materials
- Lab coat and disposable gloves
- PCR Reaction mix (8 tubes 50 ul each containing Taq DNA polymerase, MgCl2, and dNTP's)
- DNA/Primer mix (8 tubes 50 ul each containing a different DNA template with the same forward and reverse primer)
- A strip of empty PCR tubes
- Disposable pipette tips
- Cup for discarded pipette tips
- Micropipettor
- Open PCR machine
PCR Reaction Sample List
| Tube Label | PCR Reaction Sample | Patient ID |
| G16 + | Positive control | none |
| G16 - | Negative control | none |
| G16 1-1 | Patient 1, replicate 1 | 35640 |
| G16 1-2 | Patient 1, replicate 2 | 35640 |
| G16 1-3 | Patient 1, replicate 3 | 35640 |
| G16 2-1 | Patient 2, replicate 1 | 13601 |
| G16 2-2 | Patient 2, replicate 2 | 13601 |
| G16 2-3 | Patient 2, replicate 3 | 13601 |
DNA Sample Set-up Procedure
- Cut the empty PCR tubes strip in half in order to create two strips of four tubes.
- Label the sides of the tubes with the tube labels that were created.
- Set the tubes in a rack.
- The empty tube labeled as the positive control is to start first. Transfer 50 μL of the PCR reaction mix into the empty tube using a pipette.
- Using a pipette the positive control DNA needs to be transferred into the same tube as the primer. The total volume in the positive control tube should be 100 μL.
- Steps 4 and 5 need to be repeated for the negative control. Patient 1, replicates 1, 2, and 3 while patient 2, replicates 1,2, and 3. When all of this is complete, each tube should have a total volume of 100 μL.
- Secure the lids on the tubes.
- Place the tubes in the assigned PCR machine. There should be 16 slots in the heating block to put these tubes. The machine shouls not be started until each slot is filled.
Open PCR Program
- HEATED LID: 100°C
- INITIAL STEP: 95°C for 2 minutes
- NUMBER OF CYCLES: 25 - Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and extend at 72°C for 30 seconds
- FINAL STEP: 72°C for 2 minutes
- FINAL HOLD: 4°C
PCR LAB A: Research and Development
PCR - The Underlying Technology
1.) What are the different elements of the Polymerase chain reaction and what are their functions?
The template DNA acts as a reference DNA when replication of DNA begins. Primers , the location where DNA synthesis begins , are resposible for attaching to sites on the DNA strands. There must be two primers at opposite ends of the DNA segment since the primers can only go move in one direction. Deoxyribonucleotides (dNTP's) are necessary components for DNA synthesis because it allows for the taq polumerase to read the DNA code and attach matching nucleotides to creat the copies. These nucleotides essentially generate billions of DNA copies.
2.)
What happens during thermocycling and what role do the elements of the polymerase chain reaction play?
In the beginning step of the thermocycling process, the DNA is heated up at a temperature of 95 degrees celsius for two minutes. This process allows the DNA to melt enough to break the hydrogen bonds and unravel into two single stranded DNA molecules known as the process of denature. During the annealing phase, the temperature is set at 57 degrees celsius on the thermocycler for 30 seconds and the primers begin to attach to the DNA template. Then the taq polymerase comes and adds the nucleotides to the now annealed primer (72 degrees celsius for 30 seconds). The taq polymease does this all along the strand of DNA until it reaches the end of the strand and can no longer add anymore nucleotides. Finally the process is complete and the desired fragments are completed . The DNA is then cooled after all the cycles have been complete to ensure that the double helix structure of the DNA forms .
3.) Which base anneals to each? Base paring is essential to PCR . The nucleotide base adenine attaches to thymine and vise versa while Cytosine attaches to Guanine and the other way around.
| Adenine (A): | T | Thymine (T): | A | Cytosine (C): | G | Guanine (G): | C |
4.) Base-pairing occurs during the annealing and extension step of thermal cycling because during the anneal phase the primers attach to the DNA phase and base pairing is necessary for the attachment to work and during the extension step, in order for the Taq polymerase to add nucleotides to the annealed primer base paring must occur because these nucleotides are what form the structure of the DNA double helix.
LAB B:SNP Information & Primer Design
Background: About the Disease SNP
SNP stands for Single Nucleotide Polymorphisms and these are a common genetic variation within humans. Each SNP represents a single nucleotide and can replace the nucleotide (T) with (C) at anytime. SNP's are helpful in locating genes that are affected by disease. SNP's are able to predict the development of certain diseases. However, when an SNP occurs in a gene it plays a direct role in the disease of the gene, creating a negative impact on the body.
Citation: "What Are Single Nucleotide Polymorphisms (SNPs)? - Genetics Home Reference." U.S. National Library of Medicine. National Institutes of Health, n.d. Web.
Part 1: Use the NCBL database to find a disease associated sequence
What is a nucleotide?
A nucleotide is a compound consisting of a nucleoside linked to a phosphate group basic. A structural unit and building block for DNA.
What is polymorphism?
Polymorphism is a variation in the sequence of DNA among individuals.
What species is this variation found in (latin name)?
The species that this variation is found in is Homo sapiens.
What chromosome is the variation located on?
The chromosome that this variation is located on is 19:44907853.
What is listed as the Clinical Significance of this SNP?
Pathogenic is listed as the Clinical Significance of this SNP.
What condition is linked to this SNP?
The condition that is linked to this SNP is Alzheimer’s disease.
Part 2: Find the DNA sequence of SNP, and the surrounding sequence What does APOE stand for?
APOE stands for Apolipoprotein E.
What is the function of APOE? (First 3 unique terms)
The function of APOE can be described in three terms known as amyloid-beta binding, antioxidant activity, and cholesterol binding.
What is an allele?
An allele is one of two or more alternative forms of a gene that arise by mutation and are found at the same place on a chromosome.
The disease-associated allele contains what codon?
The disease-associated allele contains the codon CCG.
The numerical position of the SNP is?
The numerical position of SNP is 44907853.
Part 3: Design primers for PCR
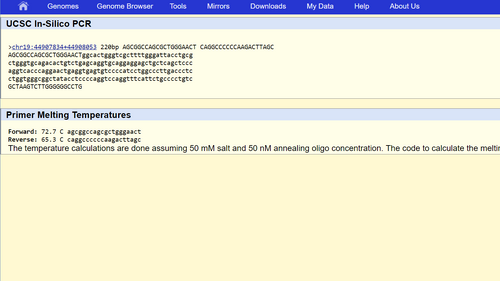
Non-disease forward primer: 5’-AGCGGCCAGCGCTGGGAACT
Numerical position 200 bases to the right of the disease SNP is: 44908053
Non-disease reverse primer: 5’-CAGGCCCCCCAAGACTTAGC
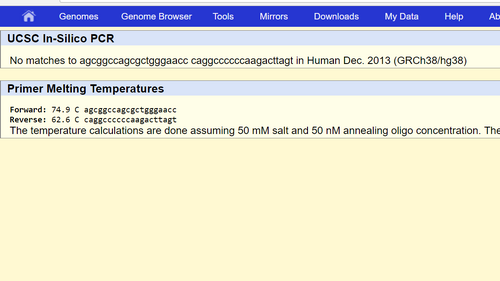
Disease forward primer: 5’-AGCGGCCAGCGCTGGGAACC
Disease reverse primer: 5’-CAGGCCCCCCAAGACTTAGT
Primer Design and Testing
When the non-disease primers were tested, the result was the following 220bp strand of DNA that started with the following letters: AGCGGCCA. When the disease primers were tested, the result was "no match" just as expected.
|}