BME100 f2017:Group15 W0800 L4
OUR TEAM
 |
 |
 |
 |
 |
LAB 4 WRITE-UP
Materials
- Lab coat and disposable gloves
- PCR reaction mix, 8 tubes, 50 μL each: Mix contains Taq DNA polymerase, MgCl2, and dNTP’s (http://www.promega.com/resources/protocols/product-information-sheets/g/gotaq-colorless -master-mix-m714-protocol/)
- DNA/ primer mix, 8 tubes, 50 μL each: Each mix contains a different template DNA. All tubes have the same forward primer and reverse primer
- A strip of empty PCR tubes
- Disposable pipette tips: only use each only once. Never reuse disposable pipette tips. If you do, the samples will become cross-contaminated
- Cup for discarded tips
- Micropipettor
- OpenPCR machine: shared by two groups
PCR Reaction Sample List
| Tube Label | PCR Reaction Sample | Patient ID |
|---|---|---|
| G15 P | Positive Control | None |
| G15 N | Negative Control | None |
| G15 1-1 | Patient 1, Replicate 1 | 96971 |
| G15 1-2 | Patient 1, Replicate 2 | 96971 |
| G15 1-3 | Patient 1, Replicate 3 | 96971 |
| G15 2-1 | Patient 2, Replicate 1 | 97071 |
| G15 2-2 | Patient 2, Replicate 2 | 97071 |
| G15 2-3 | Patient 2, Replicate 3 | 97071 |
OpenPCR Program
Heated Lid: 100°C
Initial Step: 95°C for 2 minutes
Number of Cycles: 25
- Denature at 95°C for 30 seconds
- Anneal at 57°C for 30 seconds
- Extend at 72°C for 30 seconds
Final Step: 72°C for 2 minutes
Final Hold: 4°C
Research and Development
Q1: What is the function of each component of a PCR reaction?
| Template DNA | What is being copied |
| Primers | attach to DNA and allow for replication; aka identifies which segment of the DNA the polymerase is to replicate |
| Taq Polymerase | binds to and replicates the DNA |
| Deoxyribonucleotides (dNTP's) | the free nucleotides that attach to DNA |
The Polymerase Chain Reaction function is to amplify a single section of DNA into thousands of copies of the DNA sequence. The template DNA is the section of DNA that the primers attach onto to allow for replication. The primers attach to help the Taq Polymerase locate the template DNA. The Taq Polymerase then bind to the template DNA and replicate the template strand by attaching Deoxyribonucleotides that are free in the solution.
Q2: What happens to the components (listed above) during each step of thermal cycling?
| Initial Step: 95°C for 2 minutes | the helix unwinds |
| Denature at 95°C for 30 seconds | It causes the DNA double helix to separate, resulting in two separate DNA fragments |
| Anneal at 57°C for 30 seconds | Primers attach to DNA fragments |
| Extend at 72°C for 30 seconds | Taq Polymerase attaches to the primers and begins transcription |
| Final Step: 72°C for 2 minutes | Allows time for the DNA to be replicated into |
| Final Hold: 4°C | stops PCR |
Q3: DNA is made up of 4 types of molecules called nucleotides, designated as A, T, C, G. Base-pairing, driven by hydrogen bonding, allows base pairs to stick together. Which base anneals to each base listed below?
| Adenine (A) | T | Thymine (T) | A | Cytosine (C) | G | Guanine (G) | C |
Q4: During which 2 steps of thermal cycling does base-pairing occur? Explain your answers.
PCR can be broken down into 6 steps. The first step is to raise the temperature to 95C for 2 minutes. This allows Taq polymerase to be heat activated and to be ready to bind and replicate the DNA once it is denatured. The second step is to keep the temperature at 95C for 30 seconds. This denatures the DNA which leads to two single-stranded DNA molecules. The third step is to lower the temperature to 57C for 30 seconds. This allows annealing of the primers to each of the single stranded DNA templates. Taq polymerase also binds to primer template complex and DNA formation begins. The fourth step is to raise the temperature to 72C for 30 seconds. This is the optimum temperature for Taq polymerase and allows Taq polymerase to synthesize a new DNA strand from the template strands. This process is also known as extension or elongation. It is important to note that steps 2,3, and 4 constitute a single cycle of replication. The fifth step is to keep the temperature at 72C for two minutes. This is to make sure that the remaining single-stranded DNA is fully elongated. The fifth and final step is to lower the temperature to 4C. This stops DNA replication and the PCR.
Part 1: Use the NCBI database to find a disease-associated sequence
Go to the Database of short genetic variations (dbSNP).
| What is a nucleotide | Building blocks of nucleic acids and therefore DNA |
| What is a polymorphism? | A genetic variation |
Enter rs7769452 into the search box.
| What species is this variation found in? (latin name) | Homo sapiens |
| What chromosome is the variation located in? | Chromosome 19: 44907853 |
| What is listed as the Clinical significance of this SNP? | Pathogenic |
| Click the PubMed link to view summaries of research associated with the SNP. What condition is linked to this SNP? | Alzheimer’s and subarachnoid hemorrhage |
Part 2: Find the DNA sequence of the SNP, and the surrounding sequence
Click the rs769452 link #1 search result. Look for the GeneView section.
| What does APOE stand for? | Apolipoprotein E |
| What is the function of APOE? TO find out, click the APOE link. Under Table of Contents (right side) click Gene ontology. Write the first three unique terms you see. | Amyloid beta binding, Antioxidant activity, Cholesterol binding |
Look at the Gene Model(s) section under Gene View. The non-disease allele contains the codon CTG. A change in this allele, at the T position, is linked to the disease.
| What is an allele? | Alternative form of a gene (arises from mutation) |
| The disease-associated allele contains what codon? | CCG |
What last section under Gene View shows an illustrasted sequence surrounding the SNP.
| The numerical position of the SNP is? | 44907853 |
Part 3: Now that you have a good view of the DNA sequence, you can design primers for PCR.
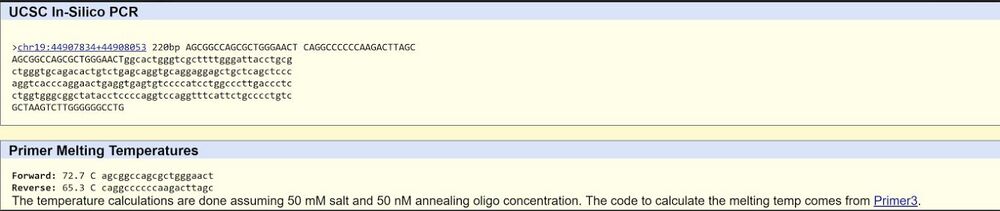
To design a non-disease forward primer, write down the sequence that is 20 bases long and ends with the nucleotide at the position you wrote in step 8.
| Non-disease forward primer (20nt) | 5’ A G C G G C C A G C G C T G G G A A C T 3’ |
Every PCR reactions needs two primers to amplify the DNA. SO, you will need to also design a non-disease reverse primer. In the sequence below, scroll 200 bases to the right of the position you recorded in step 8.
| The numerical position exactly 200 bases to the right of the disease SNP is: | 44908053 |
The reverse primer will come from the bottom (reverse) strand of DNA (bottom row of nucleotides). You will read the bottom strand from right to left this way, but write down the bases left to right.
- Write down the base at the position you wrote in the last question in the first blank space below.
- Look at the bottom sequence again. Move on base to the left and read it, and write that base in the next blank.
- Continue this process to finish the non-disease reverse primer.
| Non-disease reverse primer (20nt): | 5’ C A G G C C C C C C A A G A C T T A G C 3’ |
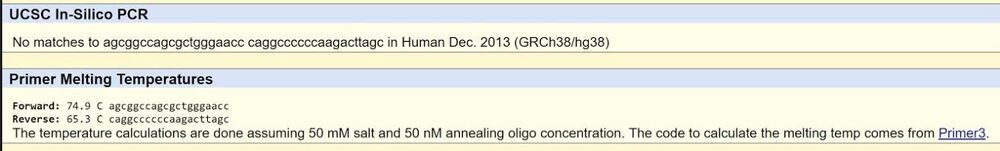
Next, design a pair of SNP-specific primers. THis part is easy. Simply change the final base of the non-disease forward primer so that it is identical to the disease SNP nucleotide, and leave everything else (including the reverse primer) the same.
| Disease forward primer (20nt): | 5’ A G C G G C C A G C G C T G G G A A C C 3’ |
| Disease reverse primer (20nt): | 5’ C A G G C C C C C C A A G A C T T A G C 3’ |
In order to understand Disease SNP, there are many other parts that have to be defined beforehand. The key pieces to specific primers are nucleotides, the building blocks of nucleic acid that essentially form DNA. However, there are instances where there happens to be a kink in the nucleotides, creating polymorphism within the DNA, resulting in a genetic variation. Disease SNP can only be found in one species, which just so happens to be Homo sapiens, and Disease SNP is found in Chromosome 19: 44907853. The reason significant reason for the labelling of Disease SNP is the fact that it is pathogenic, and is linked to both Alzheimer’s and subarachnoid hemorrhage. In addition to that, the Apolipoprotein E (APOE) is responsible amyloid beta binding, antioxidant activity, and cholesterol binding. Within the chromosomes, there are alleles which allow for alternative forms of genes, and allele shifts are responsible for mutation. The disease-associated allele contains a CCG codon, and its numerical position is 44907853.
|}