BME100 f2017:Group14 W0800 L4
TEAM MEMBERS
 |
 |
 |
 |
 |
Protocol
Materials
- Lab coat and disposable gloves
- PCR reaction mix, 8 tubes, 50 μL each: Mix contains Taq DNA polymerase, MgCl2, and dNTP’s (http://www.promega.com/resources/protocols/product-information-sheets/g/gotaq-colorless-master-mix-m714-protocol/)
- DNA/ primer mix, 8 tubes, 50 μL each: Each mix contains a different template DNA. All tubes have the same forward primer and reverse primer
- A strip of empty PCR tubes
- Disposable pipette tips: only use each only once. Never reuse disposable pipette tips. If you do, the samples will become cross-contaminated
- Cup for discarded tips
- Micropipettor
- OpenPCR machine: shared by two groups
PCR Reaction Sample List
- Patient ID Numbers
- 30003
- 67824
- Table of Samples and Labels
| Tube Label | PCR Reaction Sample | Patient ID |
|---|---|---|
| G14 P | Positive Control | none |
| G14 N | Negative Control | none |
| G14 1-1 | Patient 1, replicate 1 | |
| G14 1-2 | Patient 1, replicate 2 | |
| G14 1-3 | Patient 1, replicate 3 | |
| G14 2-1 | Patient 2, replicate 1 | |
| G14 2-2 | Patient 2, replicate 2 | |
| G14 2-3 | Patient 2, replicate 3 |
DNA Sample Set-up Procedure
- Collect all materials referenced in Materials list.
- Using separate micropipettes, collect 50 microliters of DNA/primer solution (One EACH) for Patient 1 and Patient 2 and place into test tubes.
- Label Accordingly (Patient 1: G14 1-1, Patient 2: G14 2-1).
- Using new micropipettes, collect 50 microliters of PCR reaction mix and place into test tube for Patient 1. Repeat for Patient 2.
- Run each tube through the thermocycler, using the values listed below in the "OpenPCR Program" section
- Repeat the steps above until enough samples have been collected
OpenPCR program
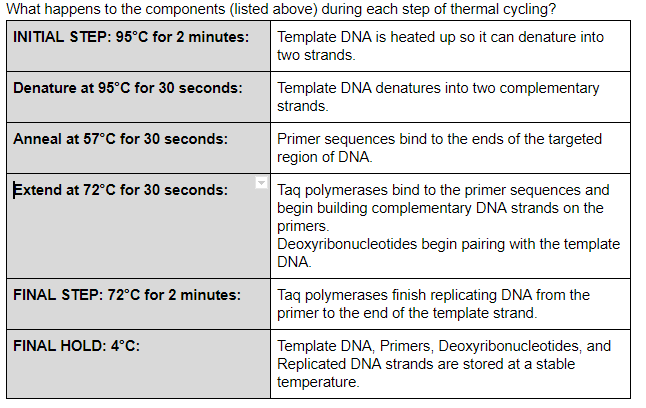
- Heated lid at 95° C for 2 minutes
- Denature at 95° C for 30 seconds
- Anneal at 57° C for 30 seconds
- Extend at 72° C for 30 seconds
- Repeat steps 2-4 for 25 cycles
- Hold at 72° C for 2 minutes
- Hold at 4° C
Research & Development
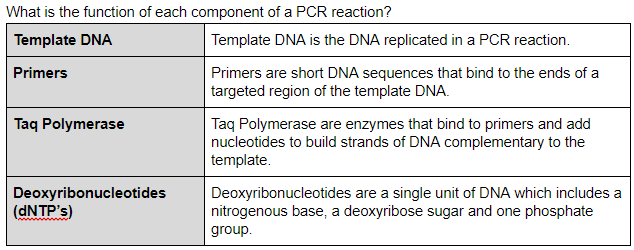
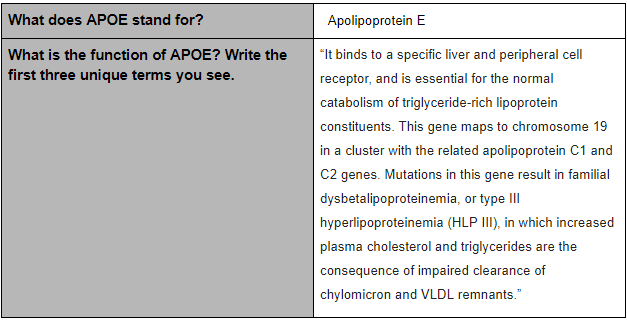
Function of Components
PCR Steps
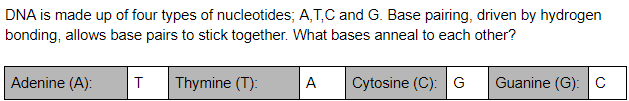
Base-Pairing
Adenine (A) and Thymine (T) bond together and Cytosine (C) and Guanine (G) bond together. Base pairing occurs during the annealing and extending stage. During the Annealing stage, primers bind to a specific location on the DNA template. These primers are complementary, meaning the nucleotides in the primer match. The next stage, Extending, is where the new DNA is made by Taq DNA polymerase enzymes. These enzymes are what add the majority of DNA bases which are paired off.
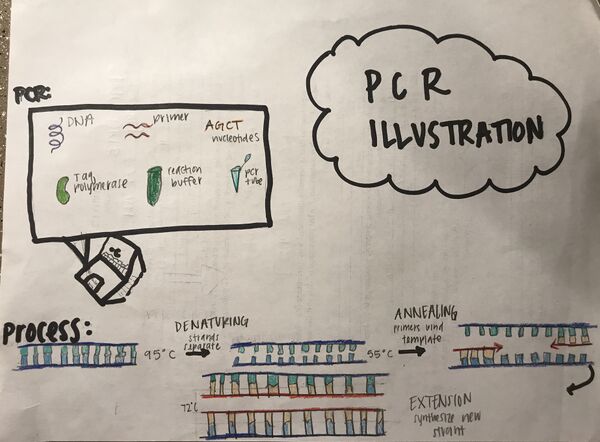
Illustration of PCR
SNP Information
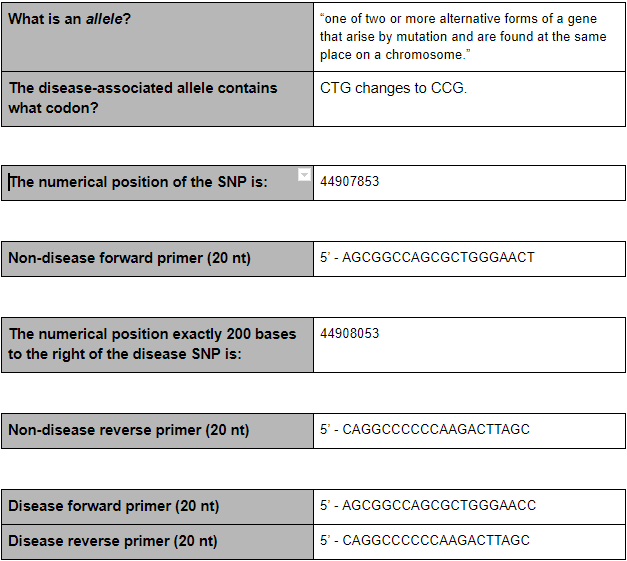
Primer Design
Summary:
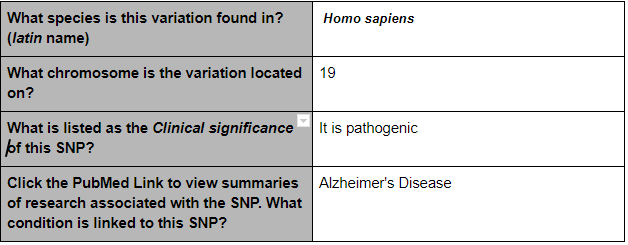
The SNP variation is found in the species of homo sapiens and on chromosome 19. If is a pathogenic and Alzheimer’s Disease is linked to this particular SNP. An allele is one of two or more alternative forms of a gene that arise from a mutation, located at the same place on a chromosome. With this disease, CTG changes to CCG.
Citations
1. Genetics Generation, “Nucleotides and Bases”, http://knowgenetics.org/nucleotides-and-bases/
2. NCBI, “Chapter 5:The Single Nucleotide Polymorphism Database (dbSNP) of Nucleotide Sequence Variation”, https://www.ncbi.nlm.nih.gov/books/NBK21088/
3. Encyclopedia Britannica, “Polymorphism”, https://www.britannica.com/science/polymorphism-biology
4. NIH, “What are Single Nucleotide Polymorphisms (SNPs)”, https://ghr.nlm.nih.gov/primer/genomicresearch/snp