BME100 f2016:Group2 W8AM L4
OUR TEAM
 |
 |
 |
 |
Materials
- Lab coat
- Safety goggles
- Disposable gloves
- CR reaction mix, 8 tubes, 50 µL each (mix contains Taq DNA polymerase, MgCl2, and dNTP’s)
- DNA primer mix, 8 tubes, 50 µL each (each mix contains a different template DNA all tubes have the same forward primer and reverse primer)
- A strip of empty PCR tubes
- Disposable pipette tips
- Cup for discarded tips
- Micropipettor
- OpenPCR machine
Patient ID's
Patient 1- 63690
Patient 2- 22568
| Tube Label | PCR Reaction Sample | Patient ID |
| G2 P | Positive Control | none |
| G2 N | Negative Control | none |
| G2 1-1 | Patient 1, replicate 1 | 63690 |
| G2 1-2 | Patient 1, replicate 2 | 63690 |
| G2 1-3 | Patient 1, replicate 3 | 63690 |
| G2 2-1 | Patient 2, replicate 1 | 22568 |
| G2 2-2 | Patient 2, replicate 2 | 22568 |
| G2 2-3 | Patient 2, replicate 3 | 22568 |
PCR Background
A Polymerase-Chain Reaction (PCR) is a technique in molecular genetics that permits the analysis of any short sequence of DNA, or RNA, and is used to reproduce or amplify selected sections of the DNA or RNA. A PCR is based on three simple steps which include:
1. Denaturation (separation) of the template into single strands.
2. Annealing (binding) of primers to each original strand for new strand synthesis.
3. Extension (copy) of the new DNA strands from the primers. These reactions may be carried out with any DNA polymerase and result in the synthesis of defined portions of the original DNA sequence.
Running a PCR and OpenPCR Program
1. Retrieve DNA sample using preferred method of extraction.
2. When running a PCR, it is suggested to keep the samples on ice. One would prepare an ice chest by putting enough ice and placing their materials inside.
3. Add the appropriate amounts of the following into a PCR tube:
- Master mix
- Forward primer
- Reverse primer
- Molecular biology grade sterile H2O
- DNA sample
4. Mix the solution by pipetting twice.
5. Put the completed PCR tube into a thermal cycler using the appropriate protocol.
During the PCR process, several things are occurring as the temperature rises and falls. The tube with the sample DNA is heated to more than 90℃ in order to separate the double-stranded DNA into two separate strands. Two primers are used; one for each strand. The tube is cooled and the primer binding occurs between 40℃-60℃. Temperature is increased to approximately 72℃. When finished, there will be two identical copies of the original DNA. The cycle begins again but this time, using the new duplicated DNA, hence the repetition of the cycles. The specific steps used in this example are shown below.
- Heated Lid - 100°C
- Number of cylces: 25
- Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and extend at 72°C for 30 seconds
- Final step: 72°C for 2 minutes
- Final hold: 4°C
Disease SNP-Specific Primer Design
- A nucleotide contains 3 parts: a base (adenine, thymine, guanine, or cytosine), pentose, and a phosphoric acid.
- A genetic polymorphism is when the phenotype of an individual is determined genetically.
- The variation specified is found in the Homo sapiens species, located in chromosome 4.
- The clinical significance of this single-nucleotide polymorphism (SNP) is that there are conflicting interpretations of the pathogenicity. Pathogenicity refers to the ability of an organism, in this case Homo sapiens, to create diseases. The condition linked to this SNP is Cardiac Arrhythmia Syndrome. During a cardiac arrhythmia, the heartbeat is irregular, in that it can be beating too fast, too slow, or in any unusual pattern.
- ANK2 stands for Ankyrin 2. Ankyrin 2 is a protein found in humans that is encoded by the ANK2 gene. When looking at the gene oncology, some of the unique terms seen were as follows: ATPase Binding, cytoskeletal adaptor activity, and enzyme binding
- An allele is an alternative form of a gene caused by a mutation. The disease-associated allele studied here contained an ATC codon.
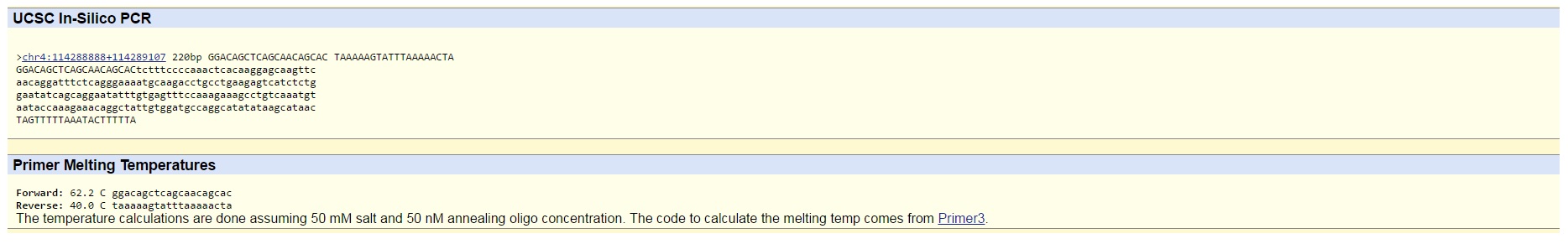
- The numerical position of this SNP is 113367751. Hence, the non-disease forward primer is GGACAGCTCAGCAACAGCAC. The numerical position of it after moving 200 base pairs to the right is 113367951. The non-disease reverse primer is TAAAAAGTATTTAAAAACTA.
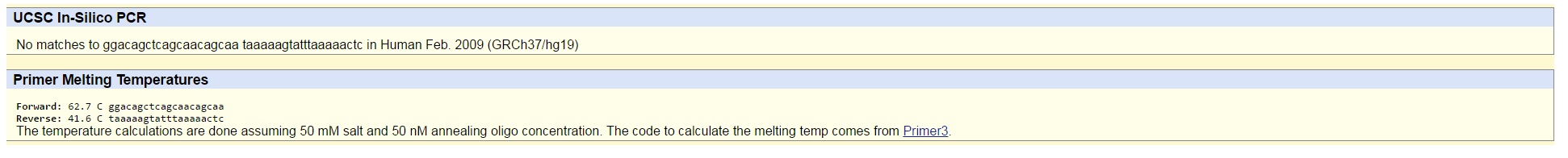
- The disease forward primer that we created is GGACAGCTCAGCAACAGCAA. We decided to exchange "C," the last letter in the non-disease forward primer, to an "A" in the disease forward primer. Accordingly, the disease reverse primer that we created is TAAAAAGTATTTAAAAACTC. We decided to exchange "A," the last letter in the non-disease reverse primer, to a "C" in the disease reverse primer. After testing the non-disease primers, the result was a 220 bp sequence from chromosome 4, hence the non-disease primers work.
 After testing the disease primers, the result was no matches.
After testing the disease primers, the result was no matches.