BME100 f2015:Group8 8amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample List
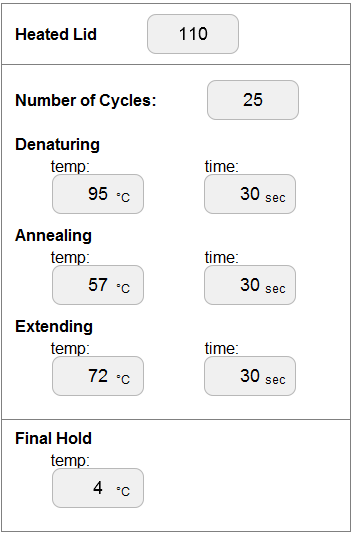
OpenPCR program
Research and DevelopmentPCR - The Underlying Technology A PCR reaction incorporates specific components to replicate DNA. We initially have a template DNA which has the specific portion of the DNA that we want to replicate. During the initial step of thermal cycling the temperature rises to 95°C were denature occurs, separating the double helix DNA into 2 separate single-standed DNA molecules. During the second step of the thermal cycling, anneal, the temperature changes to 57°C. During this portion of the PCR process, the primers attach to each end of the segment of DNA that is being copied, through a process known as base pairing. These segments are made up of specific Deoxyribonucleotides (A, T, C, and G) which have a complimentary ends. In the third step of thermal cycling, extend, temperature is brought to 72°C. At this temperature, the polymerase is activated and reads the DNA code. It then attaches the nucleotides to each of the single-stranded DNA molecules and creates two new strand of DNA. Once this step is completed, the substance is kept at 72°C for an extra three minutes to ensure that all the single stranded DNA molecules have fully extended. The substance can then be brought to 4°C to allow for short term storage.
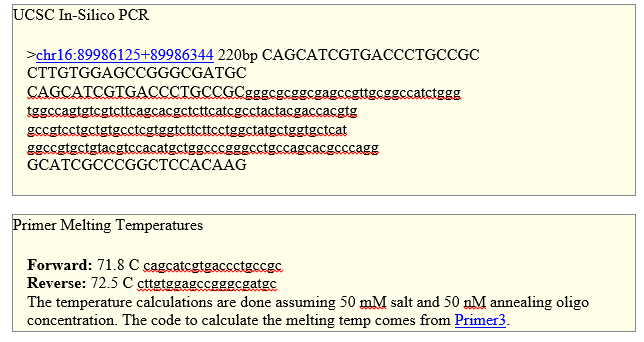
SNP Information & Primer DesignBackground: About the Disease SNP A single nucleotide polymorphism (SNP) is a single base pair difference in the nucleotide sequence between individuals of a given DNA sequence. SNP is pathogenic (disease causing). SNP is linked to being the cause of melanoma. The variation is found in Humans (Homo Sapiens). The chromosome that the variation is located on is 16:89919736 and the gene it is associated with is MC1R. MC1R stands for melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor). MC1R functions include: G-protein coupled peptide receptor activity, hormone binding and melanocortin receptor activity. The non-disease allele contains CGG. A change in this allele, at the C position, is linked to the disease. The disease-associated allele contains the sequence TGG. The numerical position of the SNP is 89919736. Primer Design and Testing To design primer pairs for PCR. Every PCR reaction needs two primers to amplify DNA. First we designed the non-disease forward primer starting by writing down the sequence that is 20 bases long and ends with the nucleotide at the numerical position of the SNP (89919736). The non-disease forward primer found was 5’- CAGCATCGTGACCCTGCCGC. Next we designed a non-disease reverse primer by first moving 200 bases to the right of the numerical position of the SNP. The numerical position exactly 200 bases to the right of the disease SNP is: 89919936. The non-disease reverse primer was determined by the reverse strand of DNA reading from right to left, writing the bases left to right. The non-disease reverse primer found was 5’- CTTGTGGAGCCGGGCGATGC. We tested the designed non-disease primers using the UCSC In-Silco PCR website. The result we got was “220 bp” sequence from the chromosome which means our non-disease primers are correct. See attached image of test. Next, we design a pair of disease SNP-specific primers by changing the final base of the non-disease forward primer so that it is identical to the disease SNP base, and leave everything else the same. Our designed forward primer is 5’- CAGCATCGTGACCCTGCCGT and our disease reverse primer is 5’- GTCGTAGCACTGGGACGGCA. We then tested the designed disease primers using the UCSC In-Silco PCR website. The result we got was “No Matches”. This indicates that current known primers that can attach to DNA strands were not able to attach. Therefore we can conclude that this is a new disease. See attached image of test.
| ||||||||||||||||||||||||||||||||||