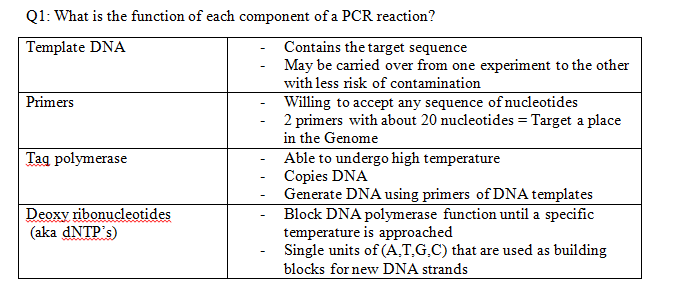
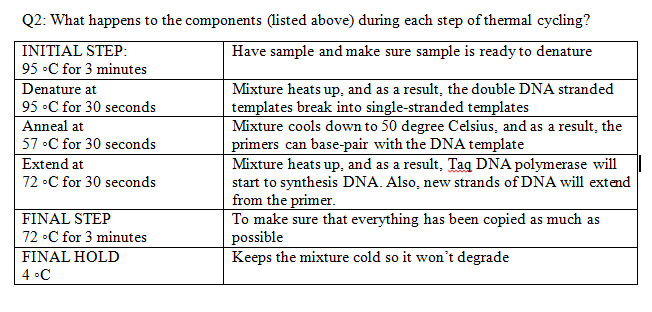
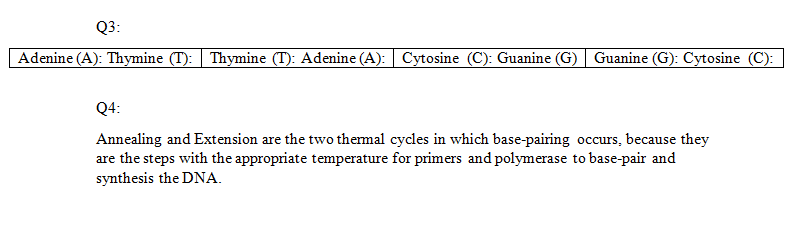
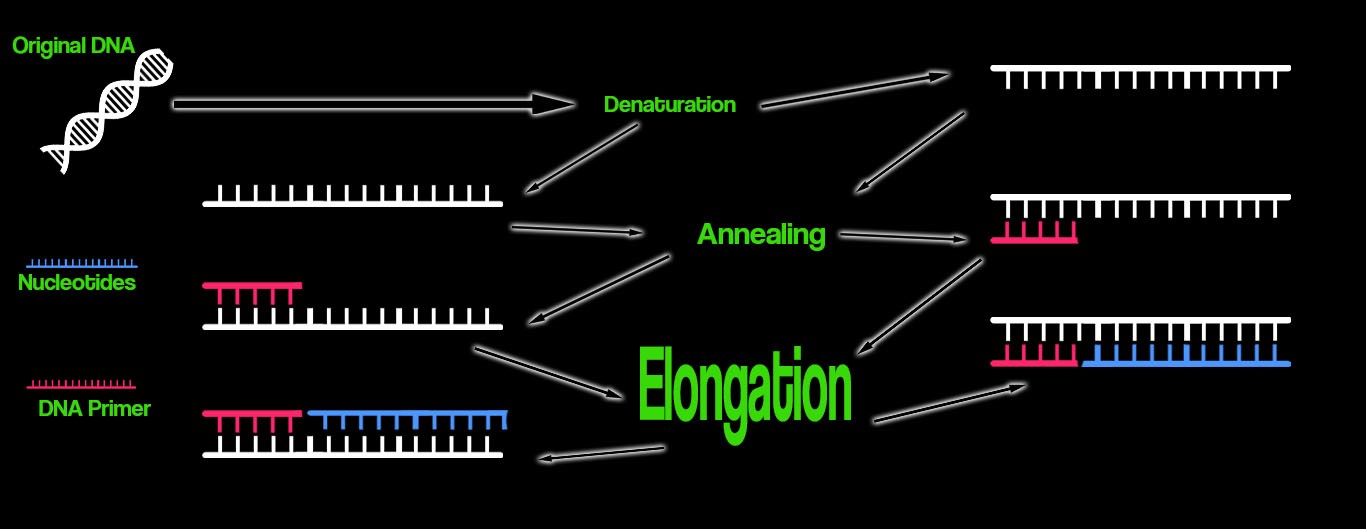
BME100 f2015:Group7 8amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||
|
OUR TEAMLAB 4 WRITE-UPProtocolMaterials
OpenPCR program
Research and DevelopmentPCR - The Underlying Technology
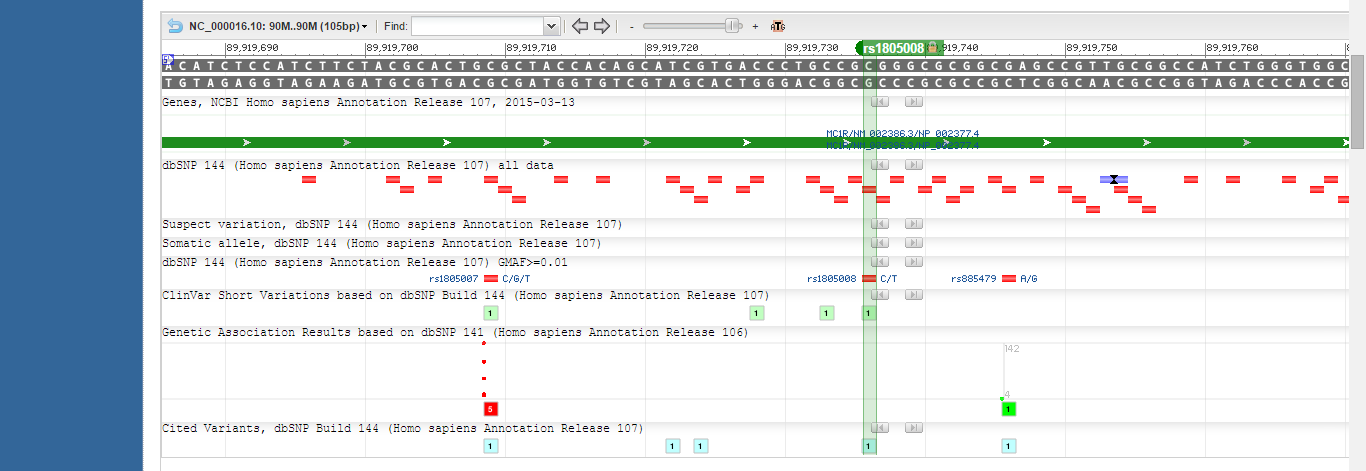
SNP Information & Primer DesignBackground: About the Disease SNP Nucleotides are the basic building blocks of RNA and DNA, and a single nucleotide polymorphism (SNP) is when a single nucleotide in a sequence is different from another sequence. Alleles are different versions of a gene, and an SNP could result in a different allele for a gene. Humans inherit two versions of every gene. The specific variation described by rs1805008 is an SNP found in Homo sapiens. The SNP is located at position 89919736 on chromosome 16. The SNP is clinically significant because it is pathogenic. Rs1805008 is associated with the Melanocortin 1 Receptor (MC1R) gene. MC1R is G protein coupled and binds to hormones known as melanocortins. MC1R regulates mammalian skin color. The SNP in the MC1R gene is associated with skin cancer.
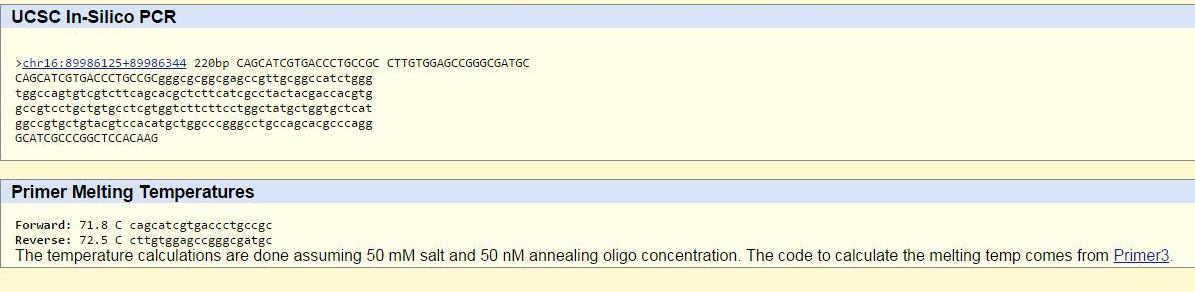
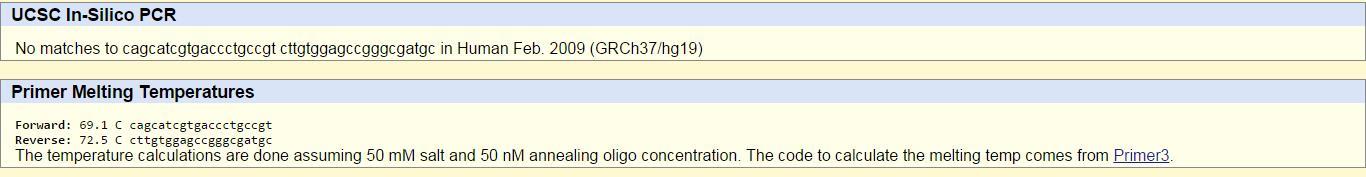
The SNP rs1805008 is located at position 89919736 on chromosome 16. In order to make a non-disease forward primer, the twenty nucleotide sequence ending in the SNP (reading from the 5’ to the 3’ end of the top strand) was taken. For the non-disease reverse primer, the twenty nucleotide sequence at the location 200 base pairs to the right of the SNP was taken (reading from the 5’ to the 3’ of the bottom strand). For the disease forward primer, the same sequence was used except thymine was substituted for cytosine in the last base pair in accordance with the SNP. The reverse primer was the same for disease and non-disease. The sequences were found using the NCBI page showing the location of rs1805008 on chromosome 16. The primers were checked on the UCSC In-Silico PCR website. The non-disease primers are correct because they will resulting in the synthesis of a 220 nucleotide sequence of DNA when they bind to the wild type sequence. The disease primers are correct because they will not bind to the wild-type sequence of DNA (they are designed to amplify the sequence with the SNP), and thus no sequence is found.
| ||||||||||||||||||||||||||||