BME100 f2015:Group6 8amL6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||
|
OUR COMPANY
LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System For this investigation, we were broken up into 17 equal teams of 6 people. There were a total of 34 patients, which means each team studied two patients each. The goal was to determine whether or not the patients were positive for the SNP. We figured this out using PCR and ImageJ to give us some data to analyze using Bayesian statistics. In order to avoid error, we made sure we had proper lab attire. For example, we wore gloves to avoid contamination of the samples. We also kept all materials under tin foil to avoid denaturing of our materials. This part, though the aim was to reduce error, may have actually also created some room for error. If one material was exposed to the light more than another, that could skew the results. It was essential that we just keep it all covered all the time. Finally, we ran 3 replicants for each patient. That way, the test was much more accurate and we had a more realistic idea of what the results should look like. Once the entire class put their results into the spreadhseet, we had our final data. There were some groups who simply did not get any results. Others tried, yet there must have been some error in the ImageJ process because their results were inconclusive. Though disappointing, this did not hinder the ability to actually run the data through Bayesian statistics using the successful trials.
Calculation 1 determines the probability a patient receives a positive final test conclusion, given a positive PCR reaction. This value is .85, or 85%. Calculation 2 determines the probability a patient receives a negative final test conclusion, given a negative PCR reaction. This value is .95, or 95%. Therefore, a patient with a negative final test conclusion will receive a more reliable PCR result than a patient with a positive final test conclusion, although for both patients the probability of a reliable PCR result is very high, since both values are close to 100%. Calculation 3 determines the probability a patient will develop the disease, clear cell renal cell carcinoma (a form of kidney cancer), given a positive final test conclusion. This value is .25, or 25%. This is the percentage, fairly low but still very significant, that a patient with the SNP sequence will end up with this form of cancer. Calculation 4 determines the probability the patient will not develop the disease given a negative final test conclusion. This value is .69, or 69%. This is the percentage, a fairly high one, that a patient without the SNP disease sequence will not develop the form of kidney cancer. This is important, because it means there is a fairly high chance, 31%, that the patient will develop clear cell renal cell carcinoma without having the SNP disease sequence test conclusion. Therefore, given Calculations 3 and 4, PCR diagnosis correlates with developing the disease, but is far from a perfectly reliable way to determine whether or not the disease will develop given a person's genome. Improper micropipetting would result in more or less original DNA sample than the device is calibrated for. Improper camera angle or distance would result in a readout of fluorimetry results that does not accurately reflect the level of amplification. Human error would also include using ImageJ in the wrong spot, which of course would amplify the wrong result. Intro to Computer-Aided DesignTinkerCAD Our Design
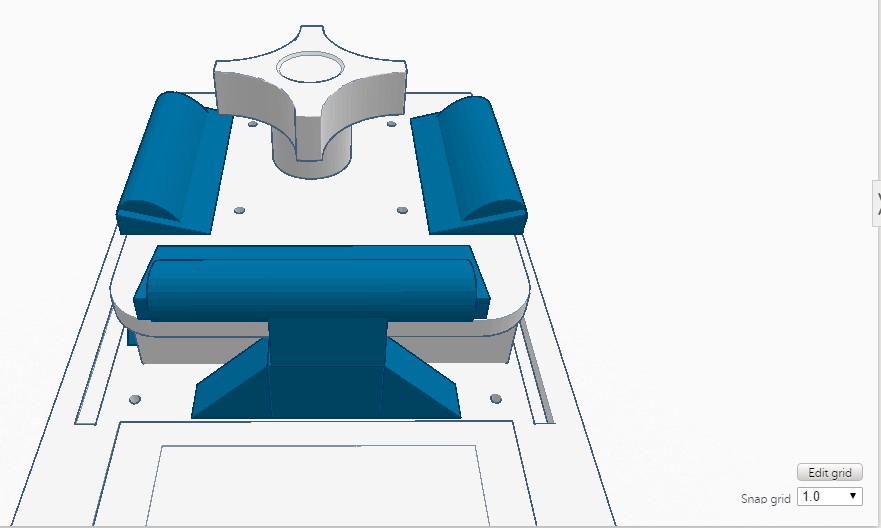
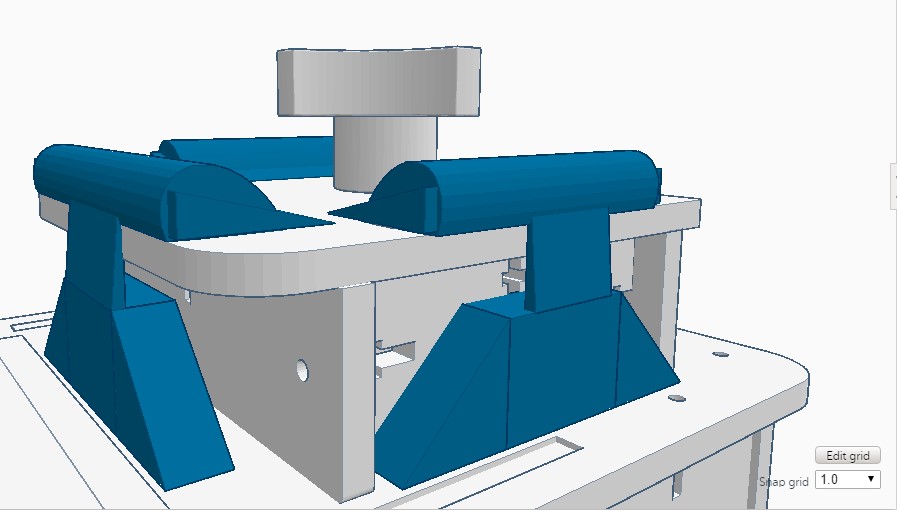
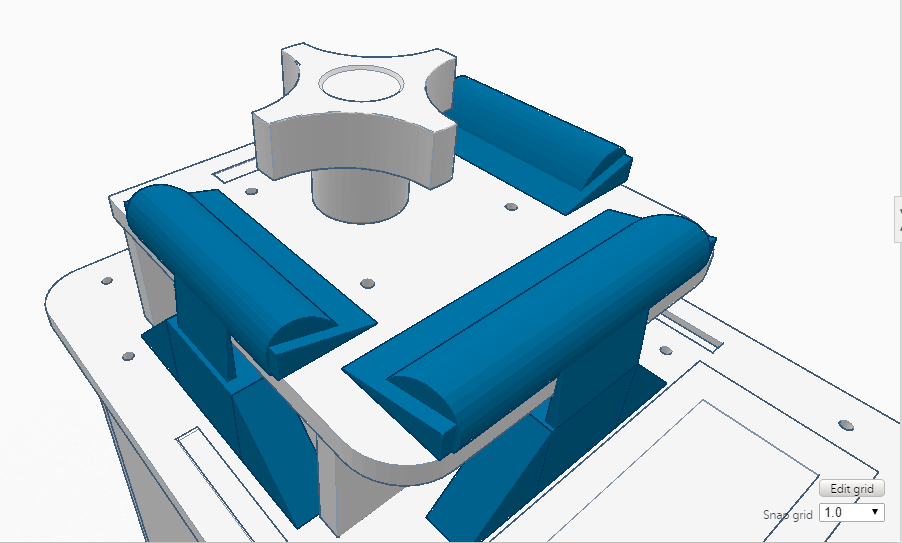
Feature 1: ConsumablesA major strength of the plastic tubes was their ability to reliably store the fluid we pipetted. One weakness of the tubes was at the bottom it was hard to whether the fluid at the bottom had thawed. This pertains to our re-designing of the lid, as the plastic tubes come into close contact with the lid. These plastic tubes, as well as the pipettor and reagents, will be included in our re-vamped PCR kit. Feature 2: Hardware - PCR Machine & FluorimeterAn important strength of the Open PCR hardware is that it is able to connect up with software to display our results, so we will want to include this in our improved Open PCR design. We will also include the fluorimeter used in the original Open PCR, because it amplifies the signal. The disadvantage of the fluorimeter is that it doesn't correct for the potential for human error, but we found this is a minor flaw compared to the flaw of how the lid operates. We will include the Open PCR machine (with a redesigned lid) as well as the fluorimeter in our system, in the same way as the original design. The only difference is that instead of a latch, the lid would be a hinge design, so it will be easier to open and close, correcting the flaw we found in Open PCR hardware.
| |||||||