BME100 f2015:Group3 8amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
Research and DevelopmentPCR - The Underlying Technology
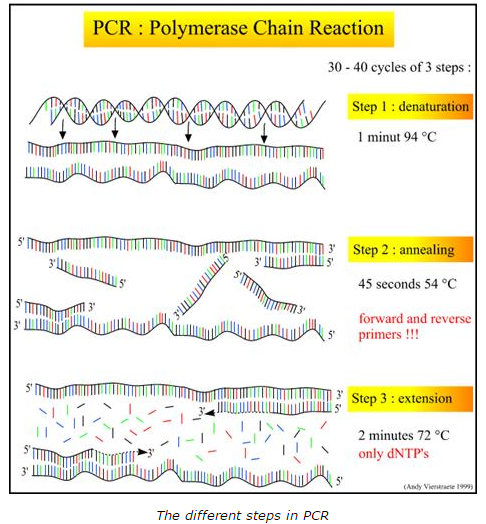
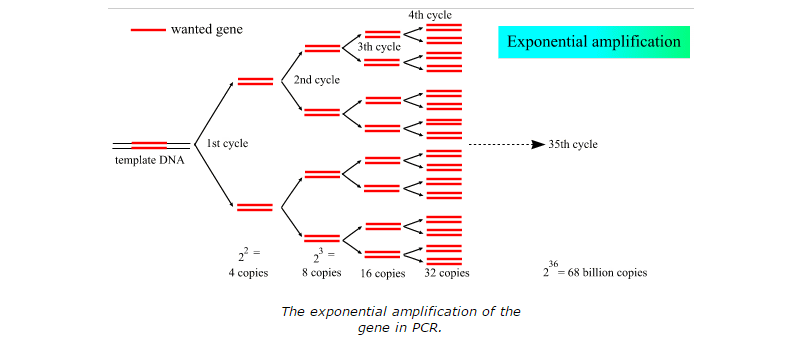
The template DNA's function is to copy a cell's DNA before it divides in two. The DNA polymerase that's most often used in PCR comes from a strain of bacteria called Thermus aquaticus that live in the hot springs of Yellowstone National Park. Primers are short pieces of DNA that are made in a laboratory and they are custom built to match to the segment of DNA you want to copy. Taq Polymerase allows the amplification of the short segments of DNA through its ability to withstand the high temperatures necessary for the PCR reaction. Deoxyribonucleotides or dNTP's allow the DNA strand to grow due to the nucleotide base they contribute. Both images attributed to: http://missinglink.ucsf.edu/lm/molecularmethods/pcr.htm
What happens to the components (listed above) during each step of thermal cycling? Initial denaturing: (95 degrees Celsius for 2 minutes) In the process of PCR, there are several steps to follow. During the initial step, the first denaturing of the template DNA occurs at a temperature of 95 degrees Celsius for 2 minutes. The Taq polymerase should be added depending on the time it took for the initial denaturation. If it takes less than 2 minutes, the Taq DNA Polymerase can be added into the initial reaction mix, otherwise it will need to be added after the first denaturation. This is because the stability of the enzyme decreases dramatically at high temperatures over 95 degrees Celsius. Denaturation: (95 degrees Celsius for 30 seconds) During the denaturing step, the DNA strands from the template DNA separate by heating to 95 degrees Celsius. The hydrogen bonds between the two strands break down forcing them to separate. Annealing: (57 degrees Celsius for 30 seconds) During the annealing process, it allows the primers to bind to their complementary DNA sequences In the DNA by forming hydrogen bonds. The annealing of the target sequences and primers is accomplished by cooling the DNA to 57 degrees Celsius. The overall time to anneal takes approximately 30 seconds. Extend: (72 degrees Celsius for 30 seconds) In the extension process The Taq DNA Polymerase binds to the template DNA and begins adding nucleotides that are complementary to the initial strand. This process is done at a temperature of 72 degrees Celsius as it is the ideal environment for Taq polymerase. Final Step: (72 degrees Celsius for 2 minutes) During this step, the samples are incubate at 72 degrees Celsius for 3 minutes to fill in the extended ends of the newly synthesized PCR product. Final Hold: (4 degrees Celsius) Lastly, the final hold in the PCR process limits the activity of Taq polymerase so as to prevent potential non-specific binding of primers and extension from these sites Which base anneals to each base listed below?
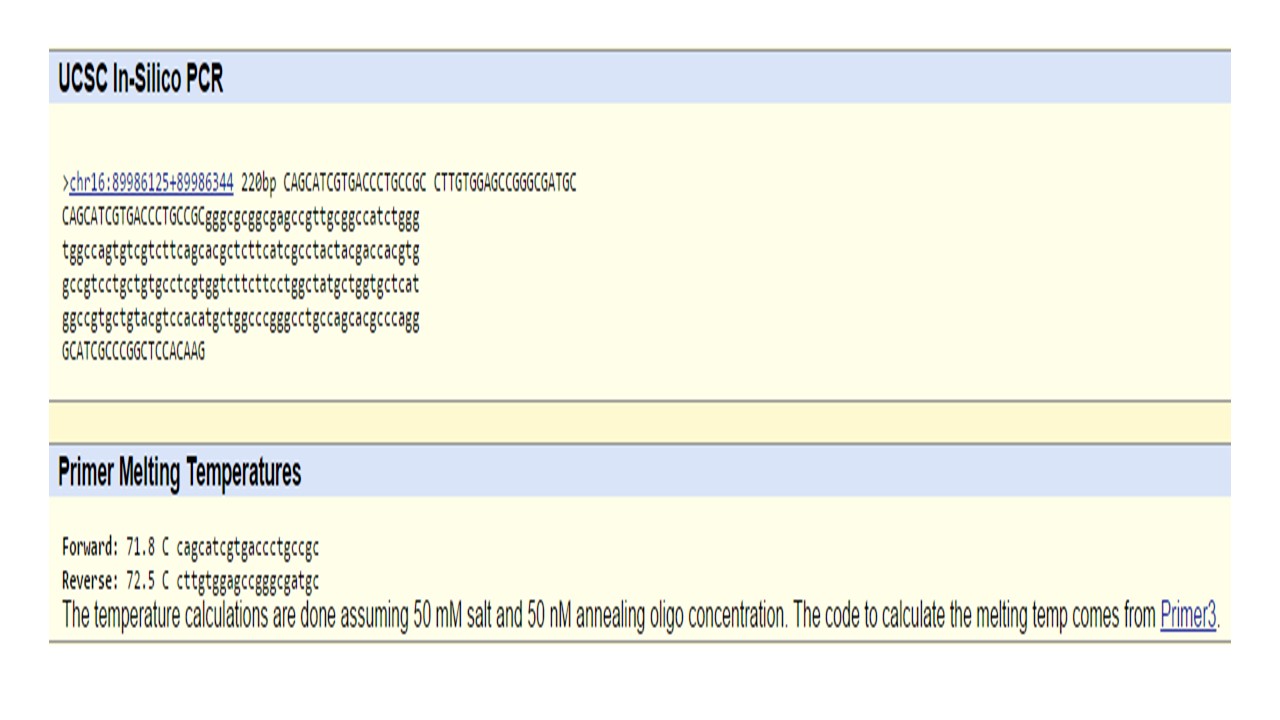
During which two steps of thermal cycling does base-pairing occur? In the steps of annealing and extension, the base pairing occurs. During the annealing process as the system is cooled, the target DNA binds with the short DNA primers that serve as guidance for the starting position for the replication to happen. When the temperature is slightly increased to 72 degrees in the extension phase, the Taq polymerase binds to the template DNA and begins adding nucleotides that are complementary to the initial strand forming the new strands of the desired DNA. SNP Information & Primer DesignBackground: About the Disease SNP A nucleotide is the basic building block of DNA. Polymorphism is the genetic variation in a species. This mutation is found in Homo Sapiens. The clinical significance of this is pathogenic. SNP is associated with the MC1R gene. Single nucleotide polymorphism is a change in a specific nucleotide. This change may or may not cause a problem for the individual with the trait. Most of the time single nucleotide polymorphism does not cause any genetic problems. While not often causing problems by themselves, they often cause vulnerabilities to diseases. Single nucleotide polymorphism is helpful by allowing people's genetic vulnerabilities to be determined. Primer Design and Testing There are two primers needed for a PCR reaction to amplify target DNA sequence. The first step in designing the primer for this experiment was to locate the non-disease allele CGG, each gene has two alleles inherited from each parent and these alleles have gene coding for the same trait, the disease is linked to the change at C position in the allele. This changed sequence associated with the disease is TGG. NCBI website was used to find the DNA sequence of the SNP and the sequence surrounding it. The numerical position of the SNP was found to be 89919736. After locating the exact numerical position of the SNP, the DNA bases could be read. Since the disease related SNP for this experiment is located on the top strand of DNA going from 5’ to 3’, the non-disease forward primer (5’ to 3’) is designed using the top strand going from left to right with a 20 base long sequence that ends at the nucleotide C which is the position of the SNP. The non-disease forward primer designed was 5’- CAGCATCGTGACCCCTGCCGC. The second primer needed for this PCR reaction is the non-disease reverse primer. To be able to design the second reverse primer the position 200 bases to the right of the position of the diseased SNP is located and that position is 89919936. The reverse primer is designed from the bottom reverse strand of the DNA. To design the primer the bottom DNA sequence is read from right to left and the bases for the reverse primer are arranged and written from left to right. The non-disease reverse primer designed was 5’-CTTGTGGAGCCGGGCGATGC. Next step for this experiment was to design a pair of disease SNP-specific primers. The whole process was the same except the final base C of the non-disease forward primer was replaced with the disease related base in this particular allele of the SNP. The disease forward primer designed was 5’-CAGCATCGTGACCCTGCCGT. The disease reverse primer designed was 5’- CTTGTGGAGCCGGGCGATGC. The UCSC In-Silico PCR website was used to check the validity of the primers. The non-disease forward and reverse primers were checked and the result came back 220 bp sequence from chromosome 16 related with this particular case indicating that the primers are valid. However, the result for the disease forward and reverse primers came back showing no matches. Since the primers were validated by using the UCSC In-Silico PCR website that only contains the non-disease human genome sequence, the match was not found for the disease specific primers. The screen shots of the results acquired are displayed below.
| ||||||||||||||||||||||||||||||||||||||||||||