BME100 f2015:Group1 8amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample List
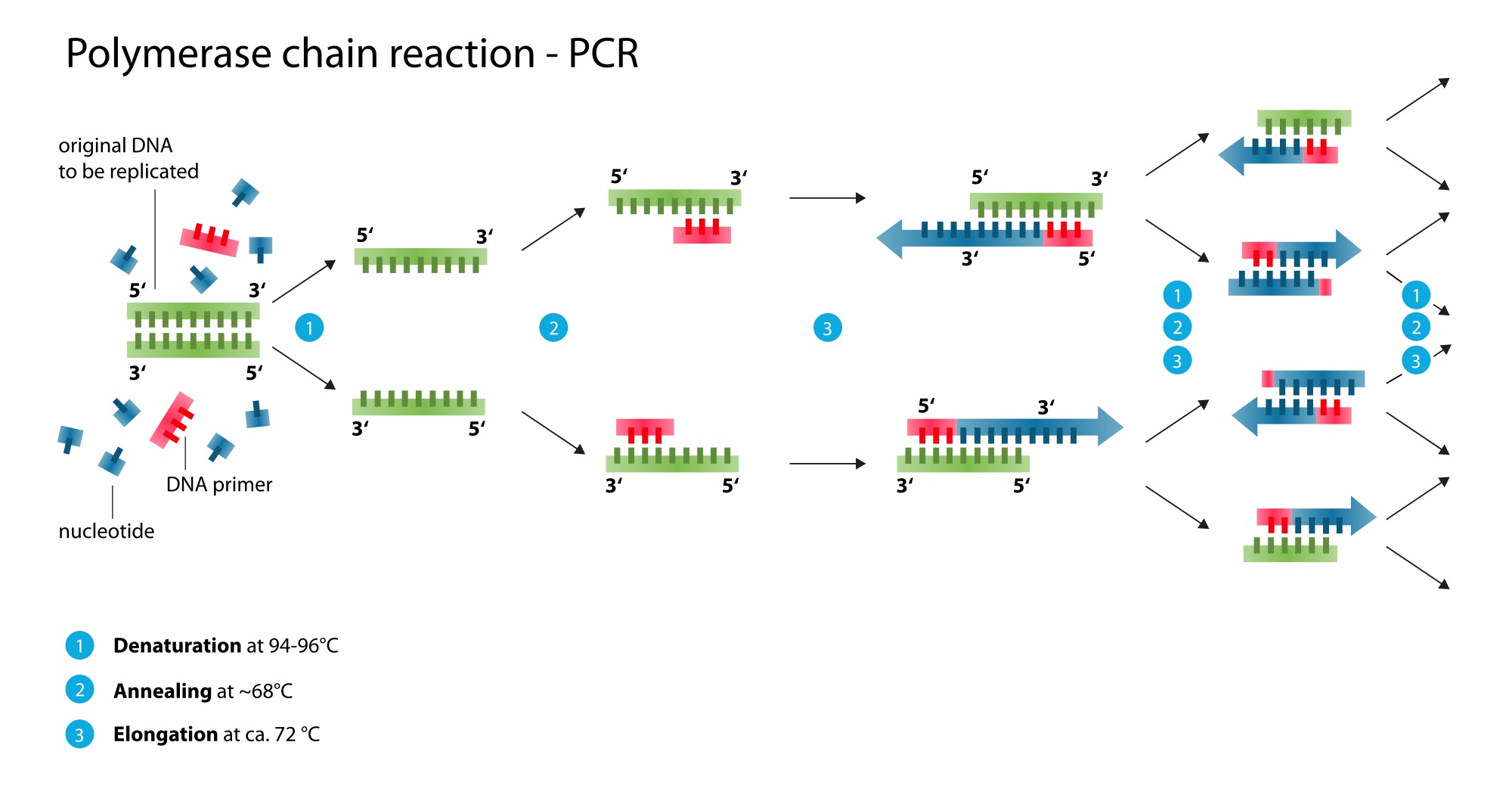
Research and DevelopmentPCR - The Underlying Technology The function of Template DNA is to be used to create a complementary strand of DNA. The function of Primers is that two primers are designed to complement the segment of DNA that is to be copied. By means of complementary base pairing, one primer will attach to the top strand at one of the segment of interest, and the other primer attaches to the bottom at the other end of the other strand. The function of Taq polymerase, is to copy a cell’s DNA before it separates into two parts. The function of Deoxyribonucleotides (dNTP’s) are to be the building blocks of new DNA strands. Question 2: What happens to the components (listed above) during each step of thermal cycling? During initial step which is at 95°C for 3 minutes, DNA double helix separates, creating two single-stranded DNA molecules. Same thing happens at step 2, temperature is raised to separate DNA strands. During denature at 95°C for 30 seconds, DNA strands split from each other. During anneal at 57°C for 30 seconds, single stranded DNA pairs try to pair up, primers will attach. In step 2, the same thing will happen, while temperature is lowered so primers will attach. Extend at 72°C for 30 seconds will activate DNA polymerase(begins to add complementary nucleotides to strand when primer is located on a single strand of DNA). It continues until it gets to the end of the strand and falls off. During the final step at 72°C for 3 minutes, Polymerase synthesizes a new dna strand complementary to the dna template strand by adding. And at the final hold at 4°C ensures that any single strains are fully extended.After the final hold from 4-15°C will be held for an indefinite time for short term storage of the reaction. Question 3: DNA is made up of four types of molecules called nucleotides, designated as A, T, C and G. Base-pairing, driven by hydrogen bonding, allows base pairs to stick together. Which base anneals to each base listed below? (A) Adenine - (T) Thymine (T) Thymine - (A) Adenine (C) Cytosine - (G) Guanine (G) Guanine- (C) Cytosine Question 4: During which two steps of thermal cycling does base-pairing occur? Explain During Anneal at 57°C for 30 seconds, where single stranded DNA pairs try to pair up, and primers will attach. And also during step two, where the process will occur again, while the temperature is lowered so that the primers will attach, and begin to add complementary nucleotides to the single strand of DNA, until it comes to the end, where the primer will then fall off.
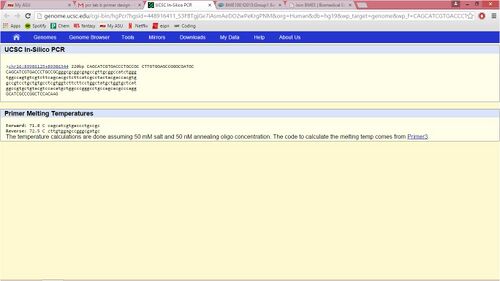
SNP Information & Primer DesignBackground: About the Disease SNP A nucleotide is a compound consisting of a nucleoside linked to a phosphate group which forms the basic structural unit of nucleic acids. A polymorphism is the presence of genetic variation within a population, upon which natural selection can operate. This variation is found in Homo Sapiens and is located in the chromosome, 16:89919736. The clinical significance of this SNP is that it is pathogenic. SNP is associated with the gene MC1R. The diseases which are linked to this SNP includes heart disease, diabetes, and cancer. Primer Design and Testing MC1R stands for melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor). Its function consists of encoding the receptor protein for melanocyte-stimulating hormone. The first three unique terms we saw were peptide, melanocortin, and ubiquitin. Allele is one of two or more alternative forms of a gene that arise by mutation and are found at the same place on a chromosome. The disease-associated allele contains the sequence TGG. The numerical position of the SNP was 89919736. The non-disease forward primer was 5'CAGCATCGTGACCCTGCCGC. The numerical position exactly 200 bases to right of the disease SNP was 89919936. The non-disease reverse primer was 5'CTTGTGGAGCCGGGCGATGC. The disease forward primer was 5'CAGCATCGTGACCCTGCCGT and the disease reverse primer was 5'CTTGTGGAGCCGGGCGATGC.
|
||||||||||||||||||||||||||||||||||