BME100 f2015:Group17 1030amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
OpenPCR program Heated Lid: 100°C Initial Step: 95°C for 2 minutes. This step allows for the hydrogen bonds in DNA to break unraveling the DNA double helix allowing primers access to the strands. Number of Cycles:25. Repeat the following Denature and Extend Steps below. Denature: at 95°C for 30 seconds; Anneal:at 57°C for 30 seconds. Annealing allows for the primers to bind with the exposed DNA. Extend: at 72°C for 30 seconds. Extending allows for DNA polymerase to start adding complementary base pairs replicating a certain gene. Final Step: 72°C for 2 minutes Final Hold: 4°C
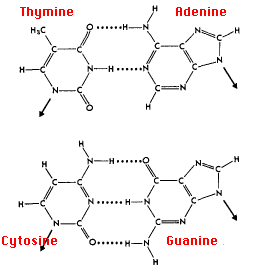
Research and DevelopmentPCR - The Underlying Technology Template DNA is used as a foundation to extract a desired section and replicate that section many times over. Two primers are used in PCR. One primer attaches to the beginning of the desired DNA site, and the other is attached to the end of the desired site when the DNA breaks up from its double helix structure. These primers help single out the sections we want to amplify. Taq Polymerase attaches to each primer site and adds complimentary base pairs to the single strand of DNA to make it double stranded. This replicates the DNA and, after a few cycles, will begin to isolate the desired section and replicate it. Deoxyribonucleotides are single units of DNA that the Taq polymerase forms to bond with a single strand of DNA in order for it to become a new strand of DNA of the replicated/desired gene. Q2
Q4. In thermal cycling, base-pairing occurs during Extending at 72º C for 30 seconds (this is where the Taq DNA polymerase is added to the single strands) and then at the Final step at 72º C for 3 minutes, (this is where the Taq DNA polymerase goes down the single stranded DNA from 3’ to 5’ adding the appropriate matches in base pairs. AT, TA, GC, and CG).
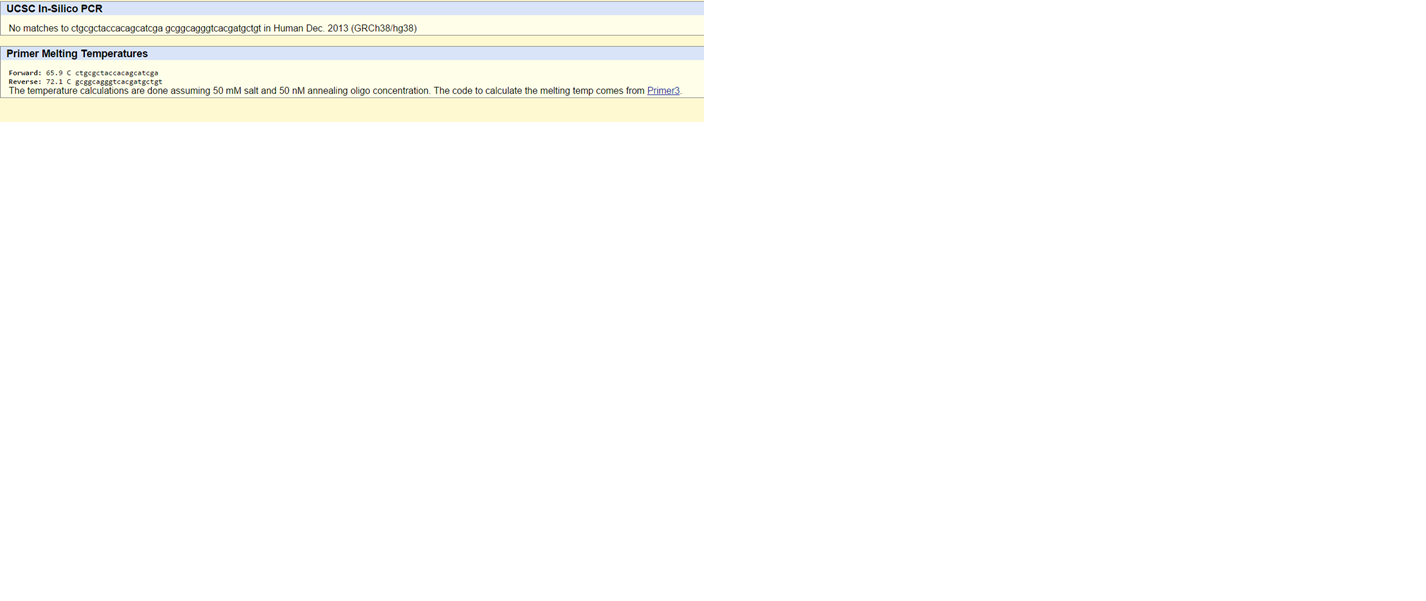
SNP Information & Primer DesignBackground: About the Disease SNP A SNP (single-nucleotide polymorphism) is a variation in DNA, based upon one nucleotide. The nucleotides in DNA are A, T, C, and G. So for example, one person's DNA could read ATGCCA and another person's DNA could read ATGCGA. While these segments of DNA do differ by a C and a G, it does not mean that the amino acid sequence for a given protein is changed. Variations are allowed, and those variations are called alleles. In most SNP's there are only two alleles. Since an SNP is a variation in the DNA, the effect could be disastrous, or have no effect at all. It just depends on the location in the DNA and if the amino acid sequence is changed. Primer Design and Testing What is a nucleotide? – The basic structural unit of nucleic acids such as DNA What is polymorphism? – The presence of genetic variation within a population What species is the variation found in? – Homo Sapiens What chromosome is variation located on? – 16 What is listed as the clinical significance of this SNP? – Other What gene(s) is the SNP associated with? – MC1R (4157) What disease is linked to this SNP? – Renal cell carcinoma, Parkinson disease What does MC1R stand for? - melanocortin 1 receptor What is the function of MC1R? – G-protein coupled peptide receptor activity, hormone binding, melanocortin receptor activity What is an allele? – One of two or more alternative forms of a gene that arise by mutation and are found at the same place on a chromosome The disease-associated allele contains what sequence? – TGG The numerical position of the SNP is – 1858 Non-disease forward primer (20nt) – CTGCGCTACCACAGCATCGT The numerical position exactly 200 bases to the right of the disease SNP: 89919936 Non-disease reverse primer (20nt) – GCGGCAGGGTCACGATGCTGT Disease forward primer (20nt) – CTGCGCTACCACAGCATCGA
| ||||||||||||||||||||||||||||||||||||||||||||||