BME100 f2015:Group15 8amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
OpenPCR program Thermal cycling is precisely alternating the heat of a solution or material from a hotter temperature to a lower temperature or from a lower temperature to a hotter temperature at specific times, through a period of time. HEATED LID: 100°C
Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and Extend at 72°C for 30 seconds
Research and DevelopmentPCR - The Underlying Technology Question 1: For this PCR reaction, DNA is extracted from cells such as hair follicles, skin or saliva. After the DNA is placed in a PCR tube, primers are added to attach to certain sites of the DNA that are wanted to be copied. Nucleotides are then added into the PCR tube to make up the DNA code and will create many duplicates of the DNA. Lastly, the DNA polymerase are then added into the PCR tube and is responsible for translating the DNA code and attaching the matching nucleotides. The Deoxyribonucleotides are used specifically because they can handle the high temperatures that the PCR reaction will create. Question 2: Once the PCR tube goes into the thermal cycler the temperature will increase to 95 degrees Celsius. During this temperature, the DNA's double helix will separate into two single stand of DNA. The temperature then drops to 50 degrees Celsius which is when the primers lock on to the target sites of the DNA before the DNA single strands try to pair up. After this portion of the cycle, the temperature will then increase to 72 degrees Celsius. At this temperature, the DNA polymerase is activated and adds nucleotides to the primers located on the strands. The DNA polymerase continues this process into it gets to the end of the DNA strand. The cycle repeats over and over again until at the final temperature of 4 degrees Celsius is reached and the target sequence has billions of copies. Question 3: Adenine (A) attaches to Thymine (T) and vise versa. The Cytosine (C) attaches to Guanine (G) and vise versa. Question 4: Base pairing happens during the step when the primers and the DNA polymerase become activated. The primers attach to the sites of the DNA before the single strands of the DNA try to pair up. When the DNA polymerase become activated, it adds nucleotides to the primers which will translate the DNA code and match nucleotides together.
SNP Information & Primer DesignBackground: About the Disease SNP A nucleotide is a compound consisting of a nucleoside linked to a phosphate group. Nucleotides form the basic structural unit of nucleic acids in DNA. A polymorphism is a DNA sequence variation occurring commonly within a population in which a single nucleotide in the genome differs between members of a biological species or paired chromosomes. This variation is found in homo sapiens. It is located on chromosome 16.The clinical significance is listed as other. It is associated with the gene MC1R. The disease most comonly associated is melanoma. MC1R stands for melanocortin 1 receptor(alpha melanocyte stimulating hormone receptor). Encodes the receptor protein for melanocyte-stimulating hormone. Is a determining factor in normal human pigment variation. An allele is one or two or more alternative forms of a gene that arise by mutation and are found at the same place on a chromosome. The disease associated allele is the sequence TGG.The position is 89919736.
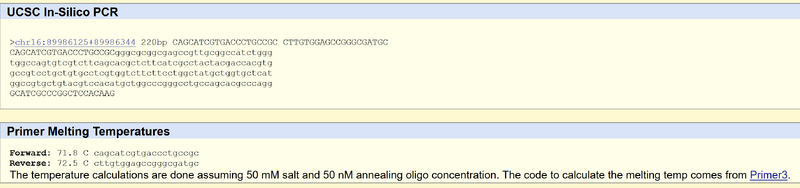
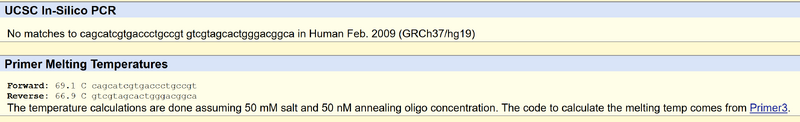
Primer Design and Testing The non-diseased forward primer starting from the five prime is 5'ACAGCATCGTGACCCTGCCGC. The Numerical position is 89919936 The Non-disease reverse primer is CTTGTGGAGCCGGGCGATGC The Disease forward primer is CAGCATCGTGACCCTGCCGT The Disease reverse primer is GTCGTAGCACTGGGACGGCA
| ||||||||||||||||||||||||||||||||||