BME100 f2015:Group13 8amL6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||
|
OUR COMPANY
| |||||||
LAB 6 WRITE-UP
Bayesian Statistics
Overview of the Original Diagnosis System
To begin this lab, our class was divided into 17 teams containing at least 6 people each. A total of 34 patients were tested by the different groups. Essentially, each group examined two patients. The main goal was to determine whether or not the patients were positive or not for the diseased SNP. The process was done using a polymerase chain reaction (PCR) to amplify the DNA of interest, and Image J to help us collect data to analyze using Bayesian statistics. In order to minimize errors, proper protective lab apparel was worn, and the patient's DNA samples were tested multiple times to give us multiple results to compare and ensure that our data was collected correctly, and accurate. However, ensuring that all the materials were under the tin foil while the PCR reaction mixture was created could have been the possible cause of error. Due to the fact that mistakes were made in always keeping the collected samples covered at all time to avoid light exposure, errors in data collection, or PCR amplification could have arisen. After all the proper precautions were taken, we started the lab. Each patients samples were tested at least 3 times so that we could ensure the tests were accurate, and so we had a better idea of what the outcome of the results should be. Once the entire class's data were collected, and results were compiled on a shared spreadsheet and assisted in the statistical analyses. Having to see every groups data was a bit challenging and unreliable because some didn't complete the lab and collect usable data. While they completed the lab their results seemed inaccurate possibly due to the errors in their ImageJ process, or other environmental errors. However, it overall didn't have that much affect on our groups results.
What Bayes Statistics Imply about This Diagnostic Approach
Calculations one and two are used to determine the accuracy of being able to detect the disease SNP. When looking at the statistics it is very clear that there is a definite correlation between the PCR replicate and the conclusion of whether a patient has the disease or not. Both the diseased and non-diseased patients came back either positive if they had the disease SNP or negative if they didn't. The results that came back both had a high correlation between PCR conclusion and disease diagnosis. This means that this method is a fairly reliable way of testing whether a patient carries the disease SNP or not.
Calculations three and four describe the possibilities of developing the disease given either a positive or negative conclusion. Both of the possibilities for each of the tests are not very appealing. Together they average around 30% for successfully predicting whether you will develop the disease. The chances of you developing the disease does come at a smaller risk, fortunately. If a positive conclusion is returned, the odds of the disease further developing is quite low. Conversely, the chances that the disease won't develop if a negative conclusion was returned is relatively high. Overall this lab doesn't really predict the development of the disease with any degree of accuracy, instead it more accurately reads if the disease is currently present.
One of the biggest hindrances of this lab was the fact that we didn't have any sort of professional way of capturing images. We had to use the cameras off of our phones that were very faulty throughout the lab. The phone camera was unable to capture the image. As the focal point of the image was within 20 cm of the camera, the phone was unable to focus. Thus, most of our pictures were not correctly in focus. In addition, the method of keeping the light out while taking the image was putting down a flap, which had drawbacks in terms of ability. We had to close it carefully or else that would shake the inside of the PCR which would alter the phone by shaking it and force it out of focus. One final possible error comes from the chance of improperly using the ImageJ program. The operators could've very easily have caused an accident by doing something as simple as improperly making a circle that didn't fit the drop. ImageJ was a tedious program that left great room for mistake. Unfortunately, we had little to no instructions to understand the functionality of the program and only followed a list of instructions as written and collected what data was produced.
Intro to Computer-Aided Design
TinkerCAD
Our company made use of TinkerCAD for improving the design of the OpenPCR. TinkerCAD is an online computer aided design software that allowed us to quickly model and adjust parameters of the original OpenPCR design. TinkerCAD was chosen over Solid Works due to the ease at which various group members could access the design online, and due to the tendency of Solid Works to often crash when saving designs. Our company used TinkerCAD to assemble the OpenPCR using components provided by the lab, and to then easily create and change any dimensions of the product. Our team chose to improve the capacity of the OpenPCR, which was easily accomplished using TinkerCAD.
Specifically, our team decided that the original OpenPCR design's limited number of microtube placements was one of the most glaring limitations of the machine. Thus, we wanted to address this issue by increasing the microtube capacity of the OpenPCR. Using TinkerCAD, we uploaded the OpenPCR shell, filled in the current holes that were designed for the lid and display. We then used the cube tool to create the correct opening size for the hinge and microtube tray and created two areas that each will hold 16 tubes for a total of 32 tubes. We also used the functionality of TinkerCAD to create a display screen for the front of the unit.
Our Design
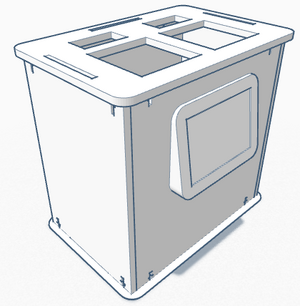
The new Open PCR design consists of two microtube trays and lids that open along the longer side of the machine. Our team believes one of the main limitation of the original OpenPCR design was the limited quantity of PCR reactions that a single machine could carry out at one time. The original design contains one tray of 16 tubes which run in a single cycle. Our design frees the machine to do multiple cycles, or a large cycle. Also, the screen was moved to the front of the machine along the side where the lids open for better visibility, and to accommodate a larger screen. The upward angled design and larger screen allows for easier viewing of the information being displayed.
Feature 1: Consumables
A major weakness in the plastic tubes used was that it was very difficult to open and close the tube without the fear of spilling the solution inside. This pertains to our re-designing of the lid, as the plastic tubes come into close contact with the lid. In addition to the redesigned OpenPCR machine, our kit will contain the redesigned plastic tubes, a micropipetter and a pack of tips, and will contain the following reagents: PCR mix, primer solution, and buffer solution. Due to the light sensitivity of SYBR Green solution, this will not be included in our consumables kit.
Consumables Included in Redesigned OpenPCR Kit
- Redesigned plastic microtubes attached in groups of 4 (108 total tubes included)
- Tube colors alternate between yellow, red, blue, and violet
- Micropipetter, 250 μL capacity
- Micropipetter tips, pack of 100
- Reagents
- PCR Mix (100 mL)
- Primer solution (100 mL)
- Buffer solution (100 mL)
Feature 2: Hardware - PCR Machine & Fluorimeter
In our system, the procedure of preparing the microtube DNA/PCR reaction mixture is the same as the original OpenPCR design. The water, buffer, dNTPs, and TAQ polymerase are added to a tube. These are added to a microtube through the use of pre-allocated micropipettor and pipette tips. The template DNA is then added to the same tube. Once this happens, the remainder of the PCR process remains the same.
The benefit of our design is that there are now 2 compartments that contain 16 open placements for tubes instead of 1 compartment. The two cycles can run independently of each other, allowing groups to run different samples simultaneously. This is an upgrade over the previous style as it allows for groups to complete the experiment at their own pace in a classroom setting such as this class. It also allows for more practical uses in the real world, as it allows for one set of samples to be created then put under the PCR process while another set of samples is created to fill the other compartment.
Our accompanying fluorimeter consists of the same set-up used in the lab with a platform to put the drops on that is back lit by a fluorescent blue light. However, in order to prevent mistakes made by the use of smart phone cameras, and to standardize focal distance, a second removable platform with a small camera will be attached adjacent to the drop. A button will be used to take the picture which can then be uploaded to a computer via a USB cord found on the second platform.





