BME100 f2015:Group12 8amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
The purpose of PCR is to replicate a known strand of DNA many times, with the purpose of yielding a higher amount of the specific DNA. Of course, a known strand of DNA is necessary for this process to occur. Initially, the known DNA has be transmitted into the specific PCR tube. The primers are then added to the same PCR tube, allowing for the DNA to be successfully copied over.If there are multiple primers, they must all be added to the tube. To actually create the DNA, nucleotides must be added to the "special tube." Finally, the addition of DNA polymerase puts all the nucleotides together to create the copied DNA. Then, the "special tube" is placed into the thermal cycler which will heat and cool the tube at specific temperatures marked in the directions above. With the corresponding temperatures marked above, the DNA is separated, bonded with primers, and creates complementary nucleotides through the help of DNA polymerase. This repeats 25 times. The result is millions of the same DNA.
Research and DevelopmentPCR - The Underlying Technology
The PCR reaction contains multiple components that each play a vital role in the replication of DNA.The first such component, and the initial component, is the template DNA. The template DNA is the strand of DNA that is being replicated. Each nucleotide in the template DNA will be assigned its base pair during the replication process, thus yielding the same copy of DNA. The next component if a primer. During the replication process, the primer attatches to the template DNA. The purpose of the primer is to act as a marker for the DNA Polymerase. The DNA starts the replication from where the primer is located on the template DNA strand. The actual creation of the DNA is done through the use of Taq Polymerase, which is a specific type of Polymerase that attaches the base-pair nucleotides together thus creating the final, replicated strand of DNA. The final component, which is also the most simple component, is the dNTP. These are the single base monomers of DNA, which usually contain a nitrogenous base, deoxyribose sugar, and a phosphate group.
To begin the process of PCR, an initial step of heating is required to calibrate the temperature of the reaction. Thus, the thermal cycler has to be heated to 95 degrees Celsius. The next step is to seperate the double stranded helix into single stranded helixes so the polymers can attach to it. The process of seperation of the double strand is called Denaturation, and this step involves heating the DNA at 95 degrees for 30 seconds. The next step, labeled as Annealing, is the attachment of polymers to the strand of DNA. As stated previously, the polymers act as a marker for the DNA polymerase to attach to and begin the nucleotide base-pair assortment. The temperature at which this occurs is 57 degrees Celsius for 30 seconds. The actual replication occurs during the Extension stage, in which the Taq Polymerase(DNA polymerase) creates a new complementary DNA strand by assorting complementary dNTPs to the already established dNTPs on the template strand. Extension is done at 72 degrees Celsius for 30 seconds. The final step, or final elongation, is the process which double checks that all single base nucleotides are paired up. This is done at 72 degrees Celsius for 3 minutes.
During the annealing process, specific base pairs are paired with one another. Adenine is bonded with Thymine, Thymine is bonded with Adenine, Cytosine is bonded with Guanine, and Guanine is bonded with Cytosine.
Base pairing occurs during the Extension step and the Final Extension step. Both steps involve the assortment of complementary dNTPs to the respective, pre-established dNTPs int he template strand. Thus, both steps involve the assortment of dNTPs in creating the overall complementary, replica of the template strand.
SNP Information & Primer DesignBackground: About the Disease SNP SNP (single-nucleotide polymorphism) occurs when a single nucleotide in the genome differs between members of a biological species or paired chromosomes (a point mutation occurs.) The specific SNP, rs1805008, is found on the MC1R-4157 gene of the 16th chromosome of Homo sapiens. This mutation is linked to increase in pheomelanin production which can lead to lightening in the skin and hair. The increase in this type of melanin is also linked to skin and UV sensitives and skin related cancers.
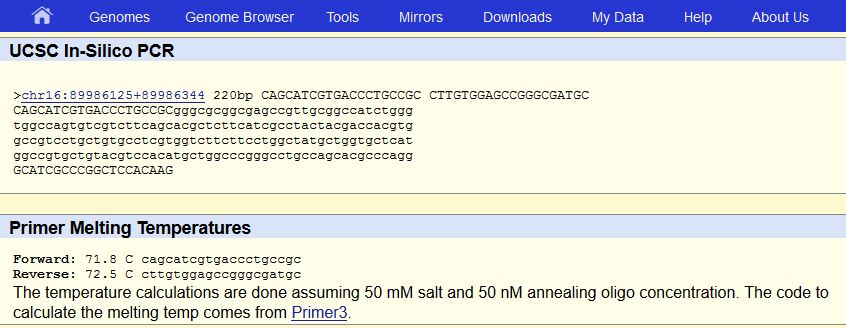
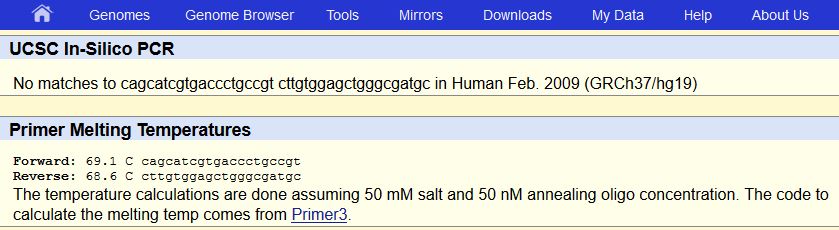
To find the DNA sequence of the SNP we used the GeneView tab on the SCBI website. Our first step was to identify the mutation we found this to be the sequence CGG changing to TGG. Next, we found the non-diseased forward primer by locating the initial position of 89919736 and reading the nucleotides from 5’ to 3’. To obtain the diseased forward primer the CGG allele had to be identified in the non-diseased forward primer then substituted with the TGG allele. Next we found the non-diseased reverse primer by adding 200 to our initial position then recording the sequence from 3’ to 5’. Then we located the CGG allele in the non-disease reverse primer and again replaced it with TGG. When we used the UCSC in-silico PCR program to check our results. We came up with a result of 220 bp for the non-diseased primers. For the diseased primers there was no match. This is most likely because the entire mutation in captured in the diseased forward primer. This wasn’t a problem when checking the non-diseased primers because there was no mutation in neither the reverse nor the forward primers. | ||||||||||||||||||||||||||||||||||