BME100 f2015:Group11 8amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
Research and DevelopmentPCR - The Underlying Technology Q1. What is the function of each component of a PCR reaction? Template DNA serves as the template for replication throughout the entire PC reaction. Primers are required to begin the process of DNA replication and are required for selective and repetitive replication. Taq polymerase serves as a copying enzyme that attaches to the template DNA and copies the strand that the primers bond to. Finally, the nucleotides are used to build the copy of the selected template DNA. Q2. What happens to the components (listed above) during each step of thermal cycling? AT 95°C the DNA double helix breaks apart into two separate strands. As the PCR tube begins to cool down the DNA strands attempt to rejoin, however, the primers stop this by bonding to their target area on each strand of DNA. Then the temperature is brought up to 72°C. When this happens the DNA polymerase becomes active and looks for primers attached to single DNA strands. The polymerase then uses the dNTP's to add complimentary nucleotides to the single DNA strand and then falls off when it reaches the end of the strand. The DNA is then held at 4°C in order to limit the activity of the Taq polymerase. Q4. During which two steps of thermal cycling does base-pairing occur? Explain your answers. The first step that base pairing occurs at is when the PCR tube begins to cool down from 95°C. At this step the primers bond onto the single strands of DNA. The second step that base pairing occurs at is when the PCR tube is heated back up to 72°C. During this step the polymerase enzyme builds replica DNA with the dNTP's.
Q3. Base Pairing
SNP Information & Primer DesignBackground: About the Disease SNP Single Nucleotide Polymorphisms is a genetic variation in a single DNA nucleotide. The rs1805008 is most commonly found in humans, and it is located in chromosome number 16. The MC1R gene, also known as melanocortin 1 receptor, determines pigmentation in skin, hair, and eye color. This gene produces two types of melanin: eumelanin and pheomelanin. Individuals that possess eumelanin have darker hair and skin, while individuals with pheomelanin tend to have lighter freckled skin. A variation in this MC1R gene increases the risk of skin cancer such as melanoma. An individual with the variation in the MC1R gene is prone to develop diseases like skin cancer, because MC1R interferes with the production of eumelanin, leading to a lack of skin pigmentation and more exposure to UV rays.
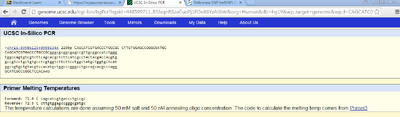
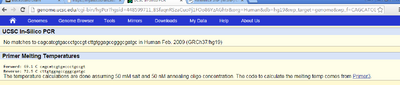
When we checked the non-disease primer in the database, and the non-disease strand came up with 220bp. The diseased primer did not find any matches. This result was to be expected because the diseased primer had a mutation in the gene; therefore, the codon would not match the non-disease (normal) sequence.To change the non-diseased primer to a diseased primer we only changed one nucleotide base from a C to a T which was on the numerical position 89919736 on the 16th chromosome, this affected the subsequent primers that would all have a changed base pair, leading to a different codon. | ||||||||||||||||||||||||||||||||||