BME100 f2015:Group10 1030amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
and dNTP’s
- These values will be used for a heating & cooling experiment on the thermal cycler. Note these steps as the experiment takes place: Heated lid: 100°C First Step to Procedure: 95°C for 2 minutes Base Number of Cycles Tested: 25 Denature at 95°C for 30 seconds, heat and allow to cooll at 57°C for 30 seconds, and Extend at 72°C for 30 seconds Final step: 72°C for 2 minutes Final hold: 4°C
Research and DevelopmentPCR - The Underlying Technology
Each of the components necessary for PCR plays an important and unique part. The template DNA is the sample DNA that contains the target sequence. The primers are short pieces of single-stranded DNA that are complementary to the target sequence and define the areas in which the target sequence are located. The taq polymerase attaches base pairs to the single stranded portions of the DNA starting at the defined primer attachment sites, while the deoxyribonucleotides are single units of the base pairs A, T, C, and G which are the essential “building blocks” for a new DNA sequence.
The steps to PCR may vary slightly according to what you are trying to do, but the basic procedure is the same. You start by heating the sample to 95 degrees celsius where the DNA strands become separated for replication by primers and polymerase. Then, you must denature the sample DNA at 95 C for 30 seconds, when the DNA double helix separates, creating two single-stranded DNA molecules. The next step is to anneal the sample at 57 C for 30 seconds to allow the two primers to bind to each single strand in preparation for replication, after which you must extend the sample at 72 C for 30 seconds to extend the activity of the primers with DNA polymerase, upon which the DNA polymerase replicates the specific strands of DNA that attached to the sample DNA. The final step is to hold the temperature at 72 C for 3 minutes, where full replication of DNA occurs and any partial copies are completed. After this, you must hold at 4 degrees Celsius where the replicated DNA is maintained until removal.
Adenine pairs to Thymine, Thymine to Adenine, Cytosine to Guanine, and Guanine to Cytosine. During thermal cycling, base-pairing occurs between the steps of annealing and extension. During annealing, the target DNA is bound with the short DNA primers that serve as starting positions for replications throughout cooling. Through the thermal cycling stage of extension, the temperature slightly increases. During this stage, two taq polymerase come in and match the base primers with their pairs, which extends the primers forming new nucleotide of target sequences of DNA.
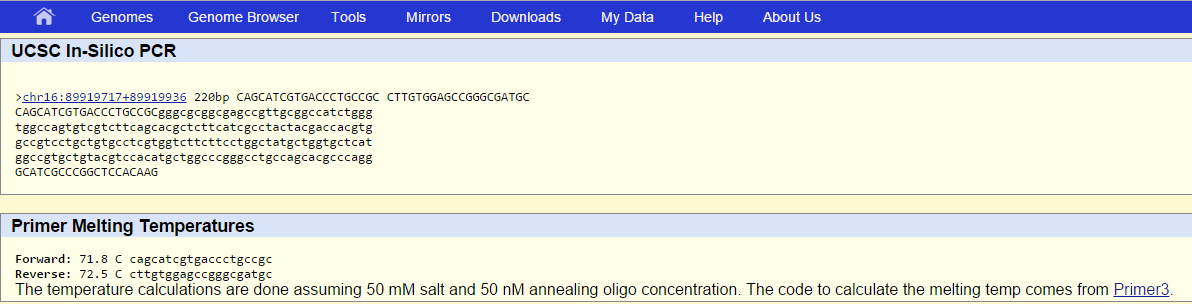
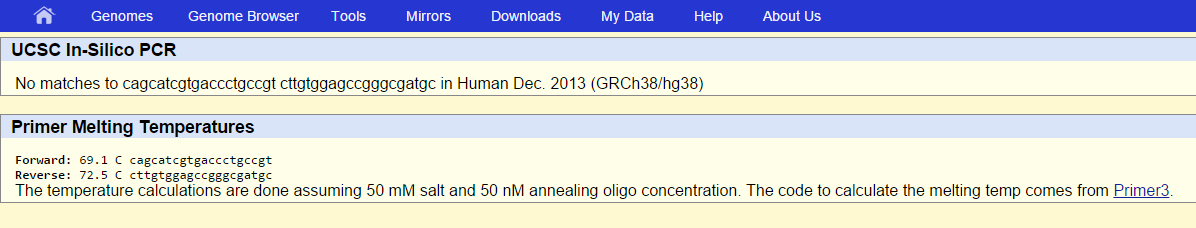
SNP Information & Primer DesignBackground: About the Disease SNP SNP stands for single nucleotide polymorphism. SNP occurs when there is a change or mutation in a sequence of nucleotides. For example, in a particular nucleotide sequence, an adenine nucleotide may be substituted with a guanine nucleotide. The gene that causes this is gene 4157. The non forward disease forward primer as determined by the experiment is 5'-C A G C A T C G T G A C C C T G C C G T and the disease associated allele is TGG. This can occur regularly in a person's genetic coding sequences and there are approximately 10 million SNP's in the human genome on average. Mutations in DNA cause this disorder. These changes can act as biological markers that help identify genes that link to SNP disorder. SNPs have many uses in the study of human health. Some include studying a person's response to certain substances and susceptibility to certain environmental factors and or diseases. Primer Design and Testing
| |||||||||||||||||||||||||||||||||