BISC110: Lab 5 Taster SNP1
Introduction
In the 1930’s Arthur Fox, a chemist at Dupont, synthesized phenylthiocarbamide (PTC). While transferring the compound to a bottle he generated a cloud of PTC dust which elicited a dramatic response in his lab mate. Although Fox tasted nothing, his colleague, C. R. Noller, complained that the dust was intensely bitter. (Arthur L. Fox’s original 1932 paper, The Relationship Between Chemical Constitution and Health, can be read at the web site of the journal, The Proceedings of the National Academy of Science, at http://www.pnas.org/content/18/1/115.full.pdf+html?sid=ef417f77-b941-4e61-ba96-a412c980cd43).
Since Fox's serendipitous discovery, variation in the ability to taste PTC (and other related bitter compounds such as those found in broccoli!) has become one of the most studied of human genetic traits. On the surface, PTC sensitivity appears to be inherited as a simple Mendelian trait with two alleles, T for taster and t for nontaster; in reality its inheritance is much more complicated. Today we know that PTC sensitivity is mediated by the PTC gene (aka TAS2R38) that encodes a bitter taste receptor (a heteromeric G-protein coupled receptor to be exact) which is found on the surface of cells of the tongue. Kim et. al.(2003), published a ground-breaking paper entitled, Positional Cloning of the Human Quantitative Trait Locus Underlying Taste Sensitivity to Phenylthiocarbamide, in the journal Science, which can be downloaded in .pdf form through the Wellesley College Library's on-line subscription to Science at http://0-www.sciencemag.org.luna.wellesley.edu/cgi/reprint/299/5610/1221.pdf). To learn even more about the PTC gene, check out the review article by Kim and Drayna (2006) at(http://chemse.oxfordjournals.org/cgi/content/full/31/7/599) and another review by S. Wooding (2006) found at (http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16636110). You should also view the video “The Science of Picky Eaters” at http://www.pbs.org/wgbh/nova/aciencenow/0404/01.html.
Additional studies of the PTC gene in humans across the globe have revealed that there are two common alleles, taster and nontaster; however, there are at least five other rare alleles that affect the taster phenotype. These different forms of the gene code for proteins that vary in their ability to bind bitter compounds such as PTC. The two common alleles of the PTC gene differ by three single nucleotide polymorophisms (SNPs) which are shown below. SNPs (pronounuced “snips”) represent the simplest type of genetic variation between individuals. A SNP refers to a specific location in the genome where different people have been shown to have a different nucleotide. If more than one percent of the population has a different base at a particular location, that location is considered a SNP. If less than one percent of the population has a different base, it is considered a mutation.
| Nucleotide position | Taster | Nontaster |
|---|---|---|
| 145 | CCA pro |
GCA ala |
| 785 | GCT ala |
GTT val |
| 886 | GTC val |
ATC ile |
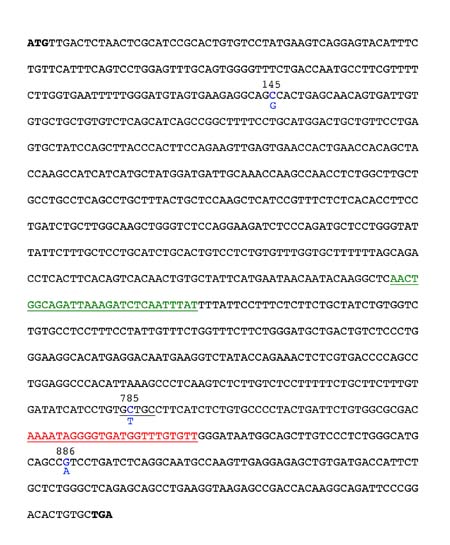
The common nontaster allele (or haplotype) has a G at nucleotide position 145 (G145), T at position 785 (T785) and A at position 886 (A886). This nontaster allele produces a polypeptide with alanine, valine and isoleucine at these sites and therefore is referred to as the AVI allele. The common taster allele has C145, C785, and G886, produces a polypeptide with proline, alanine, and valine at these sites, and is referred to as the PAV allele.
The SNP at position 785 is useful for determining your PTC genotype since the taster sequence in this region forms a Fnu4H1 restriction site while replacing C785 with T785 in the nontaster allele eliminates this restriction site. (A restriction site is a specific DNA sequence that is recognized and cleaved by a specific restriction enzyme or endonuclease.) There are two other Fnu4H1 restriction sites within the PTC gene, but we will use primers specifically designed to bind to regions flanking the one polymorphic Fnu4H1 site while excluding the other two. Using these primers in a polymerase chain reaction (PCR) generates a 303 base pair (bp) fragment. Restriction digestion of this PCR product with Fnu4H1 will yield one 303 bp fragment for the nontaster allele and two shorter fragments (65 bp and 239 bp) for the taster allele. Visit the following web site to learn more about PCR and see an animation of how it works: [dnalc.org//ddnalc/resources/pcr.html dnalc.org//ddnalc/resources/pcr.html]
Fnu4H1 Restriction Site
5’-GCNGC-3’
3’-CGNCG-5”
N can be any of the 4 bases
Figure 1. Sequence of the Taster allele of the PTC gene. The 3 SNPs are highlighted in blue with the nucleotide found in the nontaster allele shown below the taster. The position of the forward and reverse PCR primers are shown in green and red, respectively. The Fnu4H1 site you will exploit for genotyping is underlined in black at position 785.
In this three-week lab series you will determine your own phenotype and genotype at the PTC locus and explore some of the ethical issues surrounding SNPs. In the first week you will discover your PTC phenotype by performing taste tests with phenylthiocarbamide (PTC). You will also begin the process of determining your PTC genotype. This week you will extract and purify your own DNA from cheek cells and set up a PCR (polymerase chain reaction). PCR is a technique which exploits in vitro DNA replication by a heat stable DNA polymerase to produces millions of copies of a specific DNA sequence. Your PCR will amplify a region of your DNA that contains a SNP in the PTC gene. This week we will also watch a video and begin a case study in which you will explore some of the ethical issues related to the use of SNPs and other genetic information. In the second week you will determine your PTC genotype by digesting your PCR product with the restriction enzyme Fnu4H1 which cuts the taster variant of this SNP but not the nontaster variant and analyzing the products by agarose gel electrophoresis. You will also work on your case study in small groups. In the final week we will discuss the class data in preparation for writing a full lab report and each small group will present the results of their case study.
Lab 5 - Taster SNP Week 1
PART I: Taste tests
1. Taste a strip of the control paper. This will serve as a negative control and avoid possible confusion of the taste of the paper with the taste of PTC.
2. Taste a strip of the PTC paper. Record your response in your lab notebook. When we return to the lab you will record these data in class excel spreadsheet on the instructor's computer in the front of the lab. You will pick an anonymous student id number from the list available for your lab section. Record that number in your lab notebook. You will need to remember it to record your genotype results next to the phenotype results you have just obtained.
3. Enjoy a piece of candy, especially if you turn out to be a taster!
PART II: DNA extraction and purification
You will use a kit to isolate your DNA from cells which you collect from your saliva. You will chemically break these cells open and use a silica-based spin column to separate the DNA from the other cellular components.
1. Use a permanent marker to label the top of a 1.5-mL microfuge tube with a unique identifier (it need not be your initials or anything obviously identifying you), a paper cup and a 15 ml centrifuge tube with your assigned number.
2. Use the 15 ml centrifuge tube to measure ~ 10 ml of saline.
3. Pour the 10 ml of saline solution into your mouth, and vigorously rinse your cheek pockets for 30 seconds.
4. Expel the saline solution into the paper cup and transfer into the 15 ml centrifuge tube. Save the paper cup. It will serve as your liquid waste container.
WHEN YOU GET BACK IN THE LAB
5. Centrifuge the tube at 1800 x g for 5 minute to pellet your cells.
6. Decant the supernatant into the waste beaker carefully to avoid dislodging the cell pellet at the bottom. Turn the tube upside down on a paper towel to drain for a few minutes. Keep an eye on the pellet and make sure that it does not slide out!
7. Resuspend all of your cells in 400 μl of PBS (phosphate-buffered saline) by vortexing.
8. Transfer you resuspended cells to the labeled microfuge tube.
9. Add 20 μl of proteinase K to the microfuge tube.
Your lab instructor will have the tube of Proteinase K. Take your tube to them for pipetting. Proteinase K is an enzyme which lyses (breaks open) the cells and breaks down proteins. Some of the proteins broken down by proteinase K include enzymes which might otherwise break down your DNA.
10. Add 400 μl Buffer AL and immediately mix by vortexing for 15 seconds. Make sure the lid on your tube is tightly closed before vortexing.
This buffer contains a high salt concentration, which will help your DNA bind to the silica-gel-membrane of the spin column (below).
11. Incubate your tube at 56C for 10 minutes.
This incubation is to give the cells time to lyse.
12. Add 400 μl of 100% ethanol to the microfuge tube. Mix thoroughly by vortexing for 15 seconds.
13. Briefly microfuge to prevent your solution from sticking to the inside of the lid. (Press the short spin button on your microfuge.)
14. Locate a DNeasy spin column and three collection tubes. Remove 700 μl of your sample and apply to the DNeasy spin column (while the column is in one of the collection tubes). Microfuge at 8000 rpm for 1 minute.
These spin columns will only hold about 700 μl of fluid, so you will repeat this process in step 15 in order to move all your DNA onto a single spin column.
15. Remove the spin column and discard the flow-through by dumping the contents of the collection tube into your liquid waste container.
16. Place the spin column back into the same collection tube (which should now be empty). Add the remainder of your sample to the spin column. Microfuge at 8000 rpm for 1 minute.
If you have any precipitate which has formed in your original tube by this stage, pipette all of this (along with the liquid) onto the spin column. The DNA from your cells is now binding to the silica-gel-membrane in the spin column.
17. Remove the spin column and discard the flow-through into your liquid waster container and the collection tube into the autoclave bag. Place the spin column in a new, clean collection tube.
18. Open the spin column (which is sitting in its new collection tube) and add 500 μl of Buffer AW1. Microfuge at 8000 rpm for 1 minute.
19. Discard the collection tube and the flow-through. Place the column in a new collection tube.
20. Open the spin column and add 500 μl of Buffer AW2. Close the cap and microfuge at maximum speed for 3 minutes. When removing your tube from the microfuge, be careful not to let the bottom of the spin column come into contact with the flow-through.
21. Remove the spin column from the collection tube and discard the collection tube.
22. Place the spin column in a labeled microfuge tube. (Label this tube on the top and the side.) Use scissors to cut off the top of the labeled microfuge tube. Save this top.
23. Add 150 μl Buffer AE to the spin column. Be sure that you deliver Buffer AE to the membrane of the spin column but take care and don't touch the membrane with your pipet tip. Let the column sit at room temperature for 1 minute. Then, microfuge for 1 minute at 8000 rpm. The DNA will be eluted (pulled off) the membrane and be in the microfuge tube. Cap the tube with the top you had previously cut off.
Buffer AE has a very low salt concentration, which helps pull the DNA off the silica-gel-membrane of the spin column. Congratulations! You have now isolated your own DNA into a microfuge tube!
24. Store your DNA on ice until you are ready to set up the PCR reaction.
PART III: PCR of your PTC gene
Next, you will set up a polymerase chain reaction (PCR) designed to amplify the region of the PTC gene which contains the middle SNP (the one which creates a Fnu4H1 site in the taster allele). We need to use PCR so that we will have enough sample to allow us to visualize the DNA on a gel. The forward and reverse priming sites for our reactions are shown in Figure 1. In your notebook record the sequences of the forward and reverse primers. Use the convention of writing the primer sequence beginning with the 5’ end. Have your instructor check your sequences before proceeding.
1. Locate a PCR tube containing a master mix bead. Use a permanent marker to label the top of the tube with your student number.
Marvel at the size of this tube. “Master mix” is the commonly used term for this subset of the PCR components: it contains TAQ polymerase, dNTPs, and buffer. In this case, the components have been dried into a bead, so you’ll dissolve the bead into solution using your primers and DNA. Be especially careful with the PCR tube. The walls are very thin and easy to damage.
2. Pipette 20 μl of primers into your PCR tube.
This tube contains a combination of the two primers necessary to replicate both strands of your DNA. The final concentration of each primer in the PCR tube will be 0.2 μM.
3. Very carefully, pipette 5 μl of your isolated cheek cell DNA into your PCR tube.
4. Mix the contents of your PCR tube well by gently flicking the bottom of the tube. Then, vortex the tube gently (at a vortex speed of 3) until the bead has gone completely into solution. The contents of the tube should be clear.
5. Using the centrifuge located next to the PCR machine microfuge the tube briefly (5 seconds) to get all the contents of the tube to the bottom.
You will need to use adapters to microfuge these tiny tubes; the microcentrifuge next to the PCR machine is outfitted with these adaptors.
6. Follow the instructions of your lab instructor for loading your sample into the PCR machine (called a “thermal cycler”). Your instructor will tell you how to record the location of your tube in the thermal cycler.
Here is the cycle we will use for your samples:
First, there is an initial step for 10 minutes at 95C, to make sure all the DNA strands are disassociated to begin with.
This is followed by 40 cycles with the following pattern:
1 minute at 95C, for “melting” the DNA
1 minute at 55C, for reannealing the DNA
1 minute at 72C, for synthesizing new DNA
There is then a 10 minute stage at 72C, to complete whatever synthesis has been started.
Finally, the tubes are chilled at 4C until someone moves them to the freezer for longer term storage.
7. Your lab instructor will return to lab after the PCR cycles are complete and transfer your PCR reactions into the freezer for storage.
PART IV. SNPs and bioethics
1. As a class we will view a short video of a Nova scienceNOW episode that explores some of the ethical and policy issues that stem from the use of SNPs and other forms of genetic testing for medical diagnosis and assessment of genetic risk.
2. With your bench mates use the internet to explore the list of Direct-To-Consumer (DTC) genetic testing companies ([(http://www.dnapolicy.org/resources/DTCcompanieslist.pdf)]) compiled by the Genetics and Public Policy Center. Select one that your group would like to explore in detail and present to the class. Notify your lab instructor of your choice.
Assignments
1. Read the following background information on restriction enzymes found at:
(http://serc.carleton.edu/files/genomics/units/snp_restriction_enzymes.pdf)
and on gel electrophoresis at:
(http://serc.carleton.edu/files/genomics/units/snp_gel_background.pdf).
2. Read background information on SNPs in your textbook, pp. 678-684.
3. Read the following background information for our case study on ethical issues in genetic testing:
The Genetics Home Reference entries on:
direct-to-consumer genetic testing [(http://www.ghr.nlm.nih.gov/handbook/testing/directtoconsumer)]
test validity [(http://www.ghr.nlm.nih.gov/handbook/testing/validtest)]
test interpretation [(http://www.ghr.nlm.nih.gov/handbook/testing/interpretingresults)]
benefits of DTC [(http://www.ghr.nlm.nih.gov/handbook/testing/benefits)]
risks and limitations of DTC [(http://www.ghr.nlm.nih.gov/handbook/testing/riskslimitations)]
PUBLIC HEALTH: A Case Study of Personalized Medicine by S. H. Katsanis, G. Javitt, and K. Hudson,published in
Science, 4 April 2008 320: 53-54 [DOI: 10.1126/science.1156604] (in Policy Forum) found
at: (http://0-www.sciencemag.org.luna.wellesley.edu/cgi/reprint/320/5872/53.pdf)
AMA DTC genetic testing policy recommendations at: (http://www.dnapolicy.org/news.enews.article.nocategory.php?action=detail&newsletter_id=35&article_id=156)
The Answers to questions 3 and 4 will be turned in at the beginning of Lab 6. You may download a .doc file of these questions here Media:Media-Homework110Lab5.doc to use as a template for your answers.
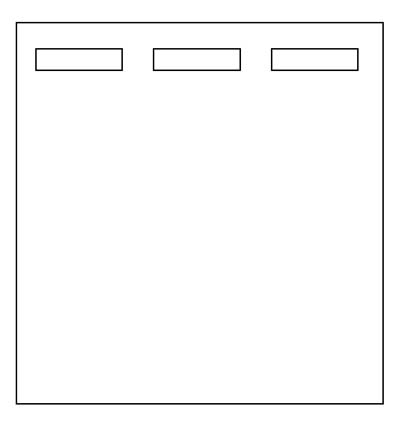
3. Think about the results you expect from next week's electrophoretic separation. After considering all you now know about PTC gene polymorphisms, draw the expected migration locations of the Fnu4H1 digested PCR product obtained from a homozygous taster (left lane), a homozygous nontaster (center lane) and a heterozygote (right lane) on a diagram of an agarose gel (template shown below).
4. Answer the following questions:
a. Define the following terms:
DTC genetic testing
analytical validity
clinical validity
clinical utility
pharmacogenetics
b. What is the American Medical Association's position on DTC?
c. Summarize the American College of Medical Genetics recommendations on DTC genetic testing.
d. List 2 potential benefits and 2 potential risks/concerns of DTC genetic testing.
e. Analytically valid tests are available for variants in the CYP450 genes. Are these tests clinically valid? Do they have clinical utility?