BIOL368/F16:HIV gp120 Exercise
This page is currently under construction.
This page is adapted from the DNA Glycosylase exercise found here.
The protein structure you will use in the exercise was published in the following article:
- Huang, C. C., Tang, M., Zhang, M. Y., Majeed, S., Montabana, E., Stanfield, R. L., ... & Kwong, P. D. (2005). Structure of a V3-containing HIV-1 gp120 core. Science, 310(5750), 1025-1028. DOI: 10.1126/science.1118398
Background
In this exercise, you will explore the structure of an HIV envelope protein necessary for infection of CD4+ lymphocytes. The exterior envelope glycoprotein, gp120, is presented as a spike on the surface of the HIV virion that is anchored to the cell via transmembrane envelope glycoprotein, gp41. Both of these glycoproteins are encoded by the HIV env gene. The binding of gp120 to the CD4 glycoprotein and, subsequently, chemokine receptors (CCR5 and CXCR4) on the lymphocyte surface enables fusion of the virion and cell membranes.
Learning objectives
We will explore the relationship between a protein’s structure and its function in HIV exterior envelope glycoprotein (gp120).
Getting started
We will begin this exercise by exploring the structure of gp120 using a molecular 3-D viewer called StarBiochem. In this particular structure, the exterior envelope protein is bound to a two-domain fragment of CD4 and an antigen-binding fragment (Fab) of a neutralizing antibody (X5) that prevents binding to chemokine receptors.
We will first focus on the structure of gp120 and then on how this protein interacts with lymphocyte proteins to accomplish membrane fusion.
- To begin using StarBiochem, please navigate to: http://mit.edu/star/biochem/.
- Click on the Start button. This will download the file
StarBiochem.jnlp. - Launch StarBiochem via the file above.
- If you receive the error message "Application Blocked by Security Settings", follow the protocol below to adjust your Java security settings:
- Open the Java Control Panel.
- Windows: Open the Control Panel. Click on Programs, then on the Java icon.
- Mac: Open System Preferences and then click on the Java icon.
- Click on the Security tab.
- Under the Exception Site List section, click on Edit Site List...
- Click Add and type in:
http://star.mit.edu. Then click Add once more and Continue in the warning window. This will allow StarBiochem to be recognized as an exception to the standard Java security protocol.
- Open the Java Control Panel.
- When presented with a security warning, check the box at the bottom of the window and click Run.
- In the top menu, click on Import > RCSB (Protein Data Bank)... In the search window, type in gp120 and then select 2B4C from the list of PDB ids. This will open the structure of "Crystal Structure of Hiv-1 Jr-fl Gp120 Core Protein Containing the Third Variable Region (V3) Complexed With CD4 and the X5 Antibody (2B4C)", which was first presented in the Huang et al. (2005) article. "2B4C" is the four character unique ID for this structure.
Take a moment to look at the structure from various angles by rotating and zooming in on the structure.
- Instructions for changing the view of a structure can be found in the top menu, under Help > Structure viewing instructions.
- The current way you are viewing the structure is by seeing each atom and bond in the protein drawn as a ball and a line, respectively. This way of representing a structure is called the ball-and-stick model and is the default model in StarBiochem. The ball-and-stick model allows you to see how atoms in the structure bond together. However, the space each atom occupies IS NOT accurately represented. To see a more realistic representation of the atoms in the structure you can use the space-filled model, where each atom is drawn as a sphere, whose size represents the physical space an atom occupies.
You can switch from the ball-and-stick model to the space-filled model in StarBiochem by increasing the size of the atoms in the structure:
- Notice that different atoms are slightly different in size.
- Gray = Carbon, Blue = Nitrogen, Red = Oxygen, Yellow = Sulfur
- Click on the Primary tab. The default atom size is 20% (ball-and-stick model).
- Move the Atoms Size slider to 100% (space-filled model).
The Reference section contains a series of terms and useful information that you will refer to during this exercise.
Exercise
- The 2B4C gp120 structure is not entirely deglycosylated. Can you differentiate between the DNA and carbohydrate components? How did you distinguish the carbohydrate from the protein?
- gp120 contains multiple sulfur atoms.
- a) Identify the name and sequence number of one of the amino acids in the structure that contains a sulfur atom.
- Using your mouse, point to a sulfur atom (yellow) in the structure. A small box appears on top of the mouse cursor indicating the name of the amino acid and its position in the amino acid sequence (ex:“[SER]156:H.HG #14367” amino acid: serine; position: 156).
- b) Is the sulfur atom located in the backbone or in the side chain of the amino acid?
- From the top menu select View -> View Specific Regions / Set Center of Rotation. This will open a smaller window, which enables you to set specific region(s) of the structure visible and centered in the viewer. Follow the steps below to view only the amino acid with the sulfur atom identified above.
- In the Protein Primary tab of the View Specific Regions / Set Center of Rotation window, select the amino acid identified above in the Sequence Window.
- Move the VDW Radius slider all the way to the left (1 Van der Waals radii). Zoom in as needed.
- First uncheck the side-chain box to hide the side-chain atoms and view only the atoms in the backbone. Next uncheck the backbone box and check the side-chain box to view only the atoms in the side-chain.
- a) Identify the name and sequence number of one of the amino acids in the structure that contains a sulfur atom.
- Next, we will explore the primary structure of the gp120 protein (Reference). The gp120 protein consists of 492 amino acids. List the 16 amino acids numbered 335 through 350 in order.
- Reset the structure by clicking Reset -> Reset structure in the top menu.
- In the Protein -> Primary tab within the main StarBiochem window, scroll down in the Sequence Window to locate amino acids 335-350 in the sequence. Refer to the Reference page for the complete name of each amino acid.
- Within a protein chain, amino acids form local structures called secondary structures (Reference).
- a) Explore the secondary structures found in gp120. Are helices, sheets or coils present in gp120? Describe the color that represents the secondary structures you observe.
- Click on the Secondary tab.
- To show the secondary structures one at a time, check the box beside the desired structure (ex: helices) and move the Structures Size slider to the right to increase the size.
- View additional secondary structures that may be present by checking the boxes next to each structure.
- b) Amino acids 335 through 350 fold into one of the secondary structures. Which secondary structure do they fold into?
- In the View Specific Regions window, go to the Protein Secondary tab and click the All button.
- Within the amino acid Sequence Window, select amino acids 335-350: click on amino acid 335, hold down Shift and click on amino acid 350.
- Move the VDW Radius slider to the left to show only the selected amino acids in the viewer.
- c) Secondary structures are held together by backbone-backbone hydrogen bonds. Name the hydrogen bond donors and acceptors that hold together the secondary structure identified above. How many residues apart are these atoms?
- In the Secondary tab, move the Hydrogen Bonds Size slider to 50.
- d) Amino acid side chains are organized in specific arrangements within secondary structures. For amino acids 335 through 350, how are the side chains oriented in 3-dimensional space in relationship to the secondary structure? Post a screenshot from StarBiochem to support your answer.
- a) Explore the secondary structures found in gp120. Are helices, sheets or coils present in gp120? Describe the color that represents the secondary structures you observe.
- Now we will explore the relationship between gp120’s structure and one of the several types of amino acids that contribute to gp120’s overall shape, its tertiary structure (Reference).
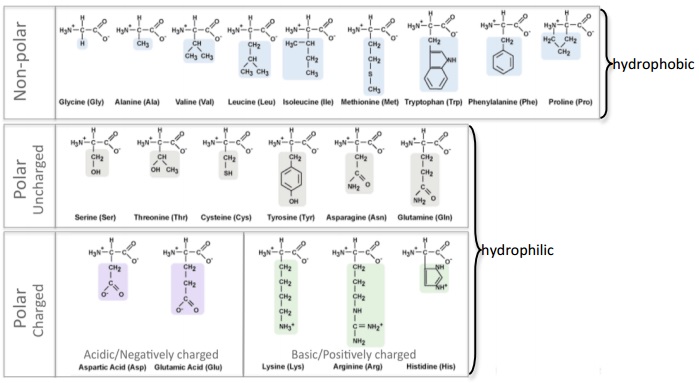
- a) Negatively charged amino acids are hydrophilic (Reference). Are the negatively charged amino acids located on the inside (buried) or outside (exposed) of this protein? What does that suggest about the cellular environment surrounding this protein, is it hydrophobic or hydrophilic? Explain your answer and provide a screenshot from StarBiochem as support.
- In the top menu, click Reset -> Reset structure.
- Click on the Tertiary tab. Move the Atoms Size slider to the left decreasing size for all the atoms in the protein.
- Click on the negatively charged/acidic button. Move the Atoms Size slider to the right to increase the atom size for the negatively charged/acidic amino acids.
- b) Charged amino acids can possess negative (acidic) or positive (basic) charge. Electrostatic attraction between acidic and basic residues can result in non-covalent interactions, which are referred to as salt bridges. Locate a salt bridge within the structure of gp120. Name the residues involved and provide a screenshot. Recall that HIV infects CD4+ lymphocytes, which can be found circulating throughout the body in the blood. How would conditions that effect blood pH such as metabolic acidosis impact the gp120 salt bridge that you identified? How could such alterations impact the structure and function of the gp120 protein as a whole?
- Click on the positively charged/basic button. Move the Atom Size slider further to the right if desired to increase the atom size for the positively charged/basic amino acids.
- c) Pairs of sulfur atoms within proteins can form covalent bonds, known as disulfide bonds. Identify a disulfide bond within the structure of gp120. What residues are involved in this interaction? Provide a screenshot from StarBiochem to support your answer. Disulfide bonds are the only covalent interactions involved in forming tertiary structure. What advantages might covalent bonding have over the other non-covalent interactions that are involved in comprising the tertiary structure of a protein?
- In the top menu, click Reset -> Reset structure.
- Click on the Tertiary tab.
- Move the Disulfide Bonds Size slider to the right to increase the size of the displayed disulfide bonds until they can be clearly visualized.
- a) Negatively charged amino acids are hydrophilic (Reference). Are the negatively charged amino acids located on the inside (buried) or outside (exposed) of this protein? What does that suggest about the cellular environment surrounding this protein, is it hydrophobic or hydrophilic? Explain your answer and provide a screenshot from StarBiochem as support.
- Now let’s explore how gp120 interacts with CD4 to recognize the CD4+ lymphocyte. In this particular structure, the gp120 protein is bound to two domains of the CD4 glycoprotein in conjunction with a two subunit neutralizing antibody, X5. Working with the different proteins present within this structure requires use of the quaternary structure (Reference) tools provided by StarBiochem. First, let’s visualize the gp120, CD4, and X5 proteins.
- Click on the Quaternary tab. Move the Surfaces Size slider to the right as needed to visualize each of the protein subunits, which will be labeled with distinct colors within a space-filled model. Overall, four unique colors should be observed throughout the structure.
- Reference page 2 of the Huang et al. paper to identify each of the protein subunits along with the V3 loop, which is part of gp120's structure.
- In this exercise, we will focus on gp120, CD4, and the V3 loop to explore how gp120 enables CD4+ lymphocyte recognition.
- a) The gp120 protein tightly binds to the CD4 glycoprotein found on the exterior of CD4+ lymphocytes. This process is facilitated by highly conserved binding cavities present in the surface of the gp120 protein. One such site is the 'Phe 43' cavity, which tightly binds the Phe43 residue present in the CD4 glycoprotein. Identify Phe43 and visualize the Phe 43 binding cavity using StarBiochem. What is the three-dimensional structure of this binding cavity? Name several of the eight gp120 residues that constitute the Phe 43 cavity. What do these amino acids have in common? Referencing section five above, what does this commonality suggest about the depth of the Phe 43 cavity? Knowing the chemical properties and structure of the Phe 43 binding cavity, how does this site accomplish tight binding to CD4 via the Phe43 residue? Provide a screenshot from StarBiochem to support your answers.
- From the top menu select View -> View Specific Regions / Set Center of Rotation. In the Protein Quaternary tab of the View Specific Regions / Set Center of Rotation window, select "chain C [Phe43]:C". Chain C refers to CD4. Then close the window used to set the center of rotation.
- In the Sequence Window, select any residue in chain G, which refers to gp120. This will highlight all amino acids present within gp120 and allow you to manipulate them as a whole.
- Move the Surfaces Translucency slider to the right as needed until the Phe43 residue from CD4 becomes visible within the gp120 protein.
- Using the Surfaces sliders, alter the appearances of CD4 and gp120 as necessary to characterize the Phe 43 cavity.
- b) gp120 mutations that impact the Phe 43 cavity, for instance at Thr257, are known to dramatically reduce the binding affinity of gp120 for CD4. Single nucleotide polymorphisms that replace Phe 43 cavity residues with what kinds of amino acids are likely to have this effect? Are viruses bearing such mutations likely to spread throughout the population?
- c) Once gp120 is bound to CD4, as in the structure we are working with, it undergoes a conformational change. The orientation of the V3 loop in this new conformation is important to proper chemokine receptor binding and membrane fusion. Looking at the gp120 structure provided by Huang et al., what role might the V3 loop play in enabling HIV to infect CD4+ lymphocytes?
- d) CCR5 is one of the chemokine receptors that can serve as an obligate second receptor for HIV to infect CD4+ lymphocytes. Certain individuals carry a rare mutation called CCR5-∆32, which involves the deletion of 32 amino acids from CCR5 receptors and renders them non-functional. What consequence may this have if such individuals are infected with HIV? Knowing that CD4+ lymphocytes differentiate from hematopoietic stem cells in bone marrow, how might a bone marrow transplant from a donor possessing the CCR5-∆32 mutation impact the life of an recipient that is at risk for contracting HIV, or that already is infected with HIV?
Reference
Primary structure
Describes the order of the amino acids in the protein chain but does not describe its shape.
Secondary structure
Describes shapes that form from local folding of regions within the amino acid chain. These smaller structures can be divided into two main types: helices and sheets. Coils are made of amino acids that do not form regular secondary structures (helices and sheets) but play important roles in protein folding.
Tertiary structure
Describes the entire folded shape of a protein chain. In addition, some proteins interact with themselves or with other proteins to form larger protein structures. These proteins have an additional level of protein structure:
Quaternary structure
Describes how multiple protein chains interact and fold to form a larger protein complex.