Arking:JCAOligoTutorial6
Sequencing Analysis
So, you've made your basic or composite part, and you think you've found a colony that contains the right product. You now want to confirm that it's right. You need to sequence to find out exactly what is in your tube. In general, sequencing is cheaper and better when outsourced to a company or core facility. So, you send out your sample, and they send you back a sequence file. In this part of the tutorial, we're going to go through how sequencing works and how to analyze the data they send you.
Before we start, you will need ApE (A Plasmid Editor) for this. So, if you haven't already done so, download it from http://www.biology.utah.edu/jorgensen/wayned/ape/. Replace the feature annotation database within ApE with an updated version. To do this, find directory in which you installed ApE on your computer and locate the file "Default_Features.txt". On my computer, it's: C:\Program Files\ApE\Accessory Files\Features. Replace this file with This Version.
How cycle sequencing works
What you really need to know
Sequencing starts with a sample plasmid DNA or PCR product and one DNA oligonucleotide. This is what you send to the sequencing facility. They do something called "cycle sequencing" to your sample, and email you back some data. You can expect between 400bp and 1000bp of "true" data that begins somewhere around 20-50 bp into the read. Where "good" and "bad" data starts and ends varies sample to sample, and we'll get into that later. The sequence you read corresponds to the region 3' of where your oligo anneals. So, you must pick the appropriate oligo to send the sequencers based on what region of the plasmid you are interested in.
Overview of the process
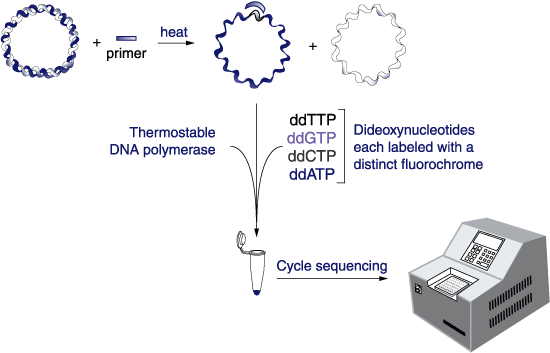
(From http:// www.biochem.arizona.edu/classes/bioc471/pages/Lecture21/AMG9.1a.gif)
The facility is going to run a reaction similar to a PCR on your sample and then load the products generated into an instrument. The reaction is going to contain many little fragments of your sequence, and the machine will separate them to single base pair resolution using capillary electrophoresis. Capillary electrophoresis is much like the gels we run in lab, but the gel is in a long narrow tube. It detects each little DNA as it comes off the column by fluorescence, and the spectrum of fluorescence (the "Chromatogram") can be interpreted by software as a string of A's, C's, T's and G's referred to as the "calls". They will send you both a text file of calls and a chromatogram file.
How cycle sequencing works
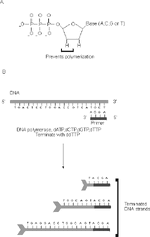
An animated description of the process is available at http://www.dnalc.org/resources/animations/cycseq.html.
From http:// www.ejbiotechnology.info/content/vol1/issue1/full/3/bip/Fig1.gif
The cycling reaction starts by denaturing your sample plasmid and annealing the oligo to its homologous sequence. The reaction is essentially a PCR reaction (dNTPs, oligo, thermostable DNA polymerase and buffer) with only one oligo, so the polymerase starts adding bases to the 3' end of the oligo. However, there are two additional components -- ddNTPs (dideoxynucleotides, in "A" of the figure) and fluorescent dyes. The dye will make the synthesized products visible in the electrophoresis instrument. The ddNTPs are chain terminators. Because they lack a 3' hydroxyl group, whenever one of these gets incorporated into a growing DNA the synthesis cannot proceed further resulting in a truncated product. For each cycling reaction, one of the 4 ddNTPs is added. So, in the reaction with ddATP, the chains get terminated at every A, and so on for all 4 reactions. The "cycling" aspect of this is that the process of denaturing, annealing, and extending is repeated so that there is linear (but not exponential) amplification of the original plasmid template.

If we were to load these cycling reactions on a normal gel like we have in lab (and they were radiolabeled, which is an even older technology called Sanger sequencing), you'd see something like the gel at left (from http:// www.cambio.co.uk/images/html_images/sequitherm_cycle1.gif). In this gel, you can see that for every vertical space on the gel there is one band from one of the lanes. You can therefore read off what base was present at each position.
In practice, since the sample is run by capillary electrophoresis, you end up with a chromatogram plotting fluorescence intensity versus time like this:
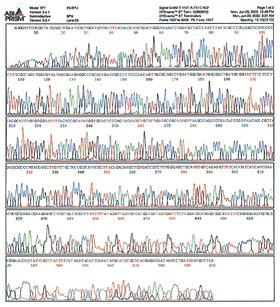
(From http:// site.hylabs.co.il/upload/infocenter/info_images/08062004105243@Sequencing-quality-.gif) That is an example of a chromatogram--the raw data from sequencing which you receive with every sample. The "calling" of the bases is just an algorithm's best interpretation of what each peak of that spectrum corresponds to. Usually the calls are pretty accurate, but occasionally the quality of the data is poor and the calls are wrong. The remainder of this tutorial deals with how you interpret the sequencing results.
Historically, the first use of dideoxy chain-terminators for sequencing DNA was the Sanger method. It involved radioactivity, so people don’t do that specific protocol anymore. However, cycle sequencing is a variation of Sanger sequencing and many people still refer to this as Sanger sequencing to distinguish it from the new multiplex sequencing methods like 454 and Illumina HiSeq sequencing which are based on a different principle. These "next-gen" methods all give shorter reads but do many thousands in parallel. They are used primarily for genome sequencing, but are also finding use now in large cloning centers. If you do a lot of cloning, "deep sequencing" solutions involving these newer technologies is a more comprehensive and cost effective approach. This is because you can pool together many experiments and sequence them in one giant read. The cost of a next-gen read is currently in the ballpark of $750. A cycle sequencing read is $2 to $10. For a lower number of samples (up to 30 for sure), it would not make sense economically nor in terms of time costs to use these technologies. Instead you want the one-sample-at-a-time method, which is an older technology.
Interpreting a sequencing result
Go ahead and download the following 2 files:
File:Jca387 ca998 2007-03-10 D02 005.txt (The calls) File:Jca387 ca998 2007-03-10 D02 005.ab1 (The chromatogram)
Open up jca387_ca998_2007-03-10_D02_005.txt in notepad, select all the text, and paste it into a window of ApE. Hit ctrl-K. This will search through the feature database and light up any features present in the sequence. Run your cursor over some of the colored text and take a look at what's in there. You've seen this plasmid before, it's pBca9145-Bca1089, the Biobricks version 2.0 RFP basic part. You should see RFP and the 4 BglBricks restriction sites: EcoRI, BglII, BamHI, and XhoI.
Let's now compare this read to the model sequence file: Media:JCASeq_pBca9145-Bca1089.str. Open up JCASeq_pBca9145-Bca1089.str in a second window of ApE. Highlight all the sequence in the sequencing read, copy it, and search for that string of text in pBca9145-Bca1089.
Uh oh...what happened? (You should have gotten an error saying "No sequence found"). Does this mean the plasmid is wrong? Um, no, not at all. In fact, this is par for the course. This is about as good as a read gets, and we know this plasmid is perfectly fine. So, what's up? Well, go ahead and launch the ab1 file into FinchTV and let's look at the raw data.
First of all, the read begins directly 3' to the spot where the oligo anneals. In this case, the oligo was ca998 (gtatcacgaggcagaatttcag), so the first few bases should have been "ataaaaaaaat". Clearly, though, the first 35 bases of this read are total garbage. That's normal. An important take-home point from this is that if your oligo anneals closer than 50bp to sequence you need to read, you're probably not going to get the data you want. From around 35bp in to around 800bp, this read looks really nice. Go ahead and select bases 35 to 813 of the sequence file in ApE and see if they match pBca9145-Bca1089. You should now be able to light up this region within pBca9145-Bca1089.
So, what can we conclude about plasmid pBca9145-Bca1089? We definitely can conclude that the part we made is absolutely correct. The quality of the read for the region between the BglII and BamHI sites is perfect here. And really, that's all we really want to get out of this sequencing effort. However, we can't say anything about the rest of the plasmid. We know nothing from this read about the sequence around the colE1 origin or the Bla gene. If we wanted to know something about those regions, we'd have to use a different oligonucleotide for sequencing that corresponded to those regions.
This first example is pretty easy. We see the entire Biobrick part, and it was a perfect match to the template. So, no worries. Now let's look at a harder-to-interpret case. Download the following:
Media:jca511_ca998_2007-05-10_F03_011.ab1 Media:jca511_ca998_2007-05-10_F03_011.txt Media:pBca9145-Bca1126.str
Put the calls file into ApE, and hit ctrl-K. Two EcoRI sites, one BglII site, and no Biobricks pop up. Now open up pBca9145-Bca1126.str and look at what should be in this file. So, is this thing wrong? Well, we can't really say anything yet, but we'll have to look at it very closely. First of all, we were only expecting one EcoRI site. The site at position 1 of the read has a good chance of being false. Open up the ab1 file and look at the region of the read. Does the chromatogram trace file support this conclusion? Looks to me like any base calls in that region are pure speculation. So, I won't worry about a potential EcoRI site.
Select bases 190-315 of the read file and search for this string of text in pBca9145-Bca1126.str. You should see that both files have this string which is a fragment of the intended Biobrick part, a phoA coding sequence. So, the thing isn't totally wrong. Select residues 16-1368 of pBca9145-Bca1126.str. You've highlighted the entire phoA part. Search for it in the calls file. It shouldn't find its cognate. Why not? First of all, notice the size of the fragment--it's 1353bp long. There is no possible way you could find the whole thing in your read. The whole file is only 1140bp long. So, you can't possibly see the entire phoA Biobrick part with one sequencing read. All we can do here is evaluate whether the sequence we do see is consistent with the model file.
Let's focus on just the N-terminus of phoA, then. Open up your trace file and let's see where the read starts to get messy. It looks pretty good until at least 650. We won't worry about any similarity between the files after 650, then. Close the trace file--we're done with it. Now find bases 400-650 in your sequence file, select them, copy them, and find that string within pBca9145-Bca1126.str. That should have worked. If it didn't, try it again. Now you have a region of brown-colored sequence selected. Copy it, search for it in the calls file, and assuming the sequence gets highlighted within the calls file, paste in the annotated version. You now should have a little patch of sequence in there that looks like this:
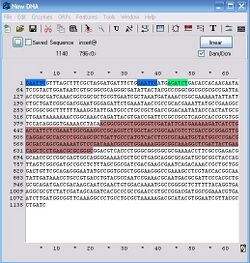
You've now marked the 3' end of the sequence you care about. Anything below that is garbage. Select the sequence within the calls file between the "true" EcoRI site and the end of the brown region (bases 34-650). If this region matches the model file, then everything we can see within this read that we care about is ok, and the clone is fine. So, copy this string and search for it in the model file. Uh oh, it doesn't match. So, there's a mutation in there somewhere. So, does that mean we're done with the analysis and can conclude this thing is a dud? No, not quite. Actually, this part is perfectly fine--it's just difficult to analyze and draw that conclusion. There is a mutation in this region, but...well keep going and you'll see what's going on.
With bases 34-650 still copied to your clipboard, go to http://pir.georgetown.edu/pirwww/search/pairwise.html and paste the sequence the sequence into the top window. Select the entire file of pBca9145-Bca1126.str into the lower window and hit enter. This program will do a pairwise alignment between the two sequences and show you where they match and where they don't. You should get something like this:

Notice the mutation? If you inspect the whole alignment, you'll see that there is only this one inconsistency in the read.
Now we know what's going on--there is one point mutation in this plasmid. Go ahead now and confirm that it's a real mutation. Select the sequence upstream of the mutation, say AGATCTGACACCACAACAATATCCGT, copy it to your clipboard, and open up the ab1 file. Search for this string within the trace file and look at the next base. Is it an A?:
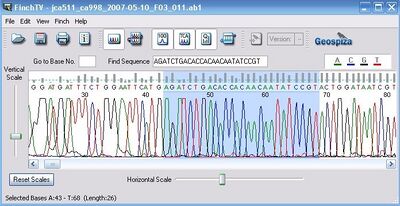
Looks like an A to me, so it's a real mutation. The next question is do we care about this mutation. The answer to that is always yes. Any mutation has the potential to be fatal, and is worthy of noting in your sequencing log and in your notebook. It doesn't necessarily mean that we throw out the plasmid.
Very often when we clone genes (from genomic DNA especially), you get mutations. The variation from the expected sequence could be an error in the pubmed file, or it could just be that the particular sample from which you obtained the genomic DNA has drifted a little from the strain the people who sequenced it originally had in their freezer. Fortunately, most (~90%) of point mutations do not alter the function of the protein product. Before we go further, let's describe what types of mutations exist:
Large Deletions and Insertions These are what the name sounds like. Either a stretch of sequence is missing or inserted into the expected sequence. This is almost always going to result in a non-functional sequence. If it shows up while cloning, something is very wrong. The plasmid is almost certainly a dud. Pick more colonies.
Frameshift mutations These are also pretty rare. It means you have a single base insertion or deletion. First and foremost, look at the trace very carefully. Often these mutations are an artifact of sequencing and aren't really present. For example, the calling program will often call a sequence like AAAAA as only 4 A's instead of 5. Repetitive bases often get miscalled. The other place this happens is within the sequence that annealed to your oligos. Remember that that region of the plasmid that specifically annealed to your oligos during construction is entirely synthetic. That sequence no longer corresponds to what was in the template sample. It's the sequence of whatever the particular oligo that got lucky and made it into the clone you grew up had. Oligo synthesis unfortunately has a fairly high rate of point deletions (meaning 1 base deleted). So, frameshift mutations where the oligos annealed happens from time to time. That's what happened, but any frameshift mutation is going to be a fatal error for a coding sequence. It will change the frame of translation, so it no longer encodes what you want.
Point mutations These are really what happen most often--single substitutions from one base to another. The most common source is polymerization error. The enzymes (Taq, Expand, Pfu, Vent, Phusion, etc.) all occasionally make point mutation errors during polymerization. The other cause is the reason described already--the template had a point mutation relative to what you expected. If you see one, it's not a good thing, but it's not necessarily fatal. There are several types of point mutations:
Nonsense mutations This means that the mutation you made, when the part is translated in its relevant frame, introduced an in-frame stop codon. This is fairly rare during cloning simply because of all the possible point mutations that could happen, the nonsense errors are only a small fraction. It is fatal, though.
Missense mutations This means that the point mutation changes the codon containing it from one amino acid to another. That mutation could be neutral, meaning it doesn't affect the protein product, or not. Most missense mutations in fact are neutral (~90%). You have no way of knowing whether your particular mutation is neutral, though, so missense mutations get hairy. So, if you get a missense mutation, you have to establish whether the error is presence in your template. If it was there, was the template construct functional? If it was, the mutation is probably neutral. If it wasn't in the template, find another clone that doesn't have the mutation. In fact, the best thing to do if you get a missense mutation in a read is to compare it to another clone. If both reads have the missense mutation, there is a high probability that the mutation was present in the template. If only one has it, most likely the mutation occurred during PCR, and you should use a different clone.
Silent mutations This means there's a point mutation, but it doesn't change the translation. These occur because of the degeneracy of the genetic code. Usually it means the mutation is at the 3rd position of a codon, which is where most of the degeneracy lies. These mutations almost never change the function of the part. If you had two clones, and one matched the expected sequence and one had a silent mutation, you'd certainly choose the one that had no errors. However, the silent mutations are ok if you can't easily find a perfect clone. Just note that there was a silent mutation, and make a new model file that reflects the sequence present in your read.
So, what kind of mutation is present in our phoA plasmid? To figure this out, you need to predict the translation of the phoA in the read and the phoA in the model file. Are the translations the same? If so, it's silent. If not, it's missense or nonsense. You should have used the translation tools within ApE in the previous exercise, so I leave it to you to figure this out, but I'll give you a hint--this one is silent, and the mutation was present in the template sample which was entirely functional. So, this guy is ok. The course of action here is to note that there was a silent mutation and modify the model file, and move on. However, since we only confirmed the N-terminus of PhoA with this read, we'll want to do another sequencing reaction over the C-terminus and make sure it's ok.
Sequencing Quiz
I've supplied a list of model files and the sequencing reads and chromatograms below. For each one, analyze the data and determine which one of the following scenarios best describes the part contained within your sample.
***Warning*** I rarely get perfect responses to this quiz...it's intentionally tricky! For all instances here, we only care about the 'part' itself--so, any sequence 5' of the EcoRI site or 3' of the XhoI sites (for BglBricks) or 5' of the EcoRI or 3' of the PstI site (for XbaI/SpeI standard) in these reads can be ignored.
Answer Choices
(A) Perfect. The clone is perfect. The entire part was visible in the read and the clone is correct. (B) Deletion/Insertion/Other Sequence. The read quality is fine, but the clone has a deletion, insertion, or no match at all to the model. What's wrong with the clone? (if it just has a point mutation, the answer is E instead.) (C) Perfect Partial. The part may be ok, but I couldn't see all regions of the part. What bases of the model file can be concluded to be "good" based on the data you saw? (Note: if the clone contains a mutation within the region you successfully read, the answer is "B" or "E".) (D) Bad read. The read quality is really bad. I can't say anything about the clone. (E) Point Mutation. The read is mostly consistent with the model file, but there's a point mutation. What kind of mutation? Should I throw out the clone based on this data?
| Model File | Oligo Used | Sample Name | Calls File | Trace File | ||
| Clone 1 | Media:JCAseq_pSB1A2-Bca9143.str | G00101 | jca388 | Media:jca388_G00101_2007-03-10_E02_004.txt | Media:jca388_G00101_2007-03-10_E02_004.ab1 | |
| Clone 2 | Media:JCAseq_pSB1A2-Bca9141.str | ca998 | jca384 | Media:jca384_ca998_2007-03-06_F03_011.txt | Media:jca384_ca998_2007-03-06_F03_011.ab1 | |
| Clone 3 | Media:JCAseq_pSB1A2-I13521.str | ca998 | jca453 | Media:jca453_ca998_2007-03-27_A01_008.txt | Media:jca453_ca998_2007-03-27_A01_008.ab1 | |
| Clone 4 | Media:JCAseq_pBca9145-Bca2048.str | ca998 | jca387 | Media:jca387_ca998_2007-03-10_D02_005.txt | Media:jca387_ca998_2007-03-10_D02_005.ab1 | |
| Clone 5 | Media:JCAseq_pSB1A2-Bca9158.str | ca998 | jca393 | Media:jca393_ca998_2007-03-16_G09_034.txt | Media:jca393_ca998_2007-03-16_G09_034.ab1 | |
| Clone 6 | Media:JCAseq_pBca1066-Bca1068.str | ca886F | jca380 | Media:jca380_ca886F_2007-02-13_G02_002.txt | Media:jca380_ca886F_2007-02-13_G02_002.ab1 |
Oligo Sequence ca998 gtatcacgaggcagaatttcag G00101 attaccgcctttgagtgagc ca886F ggtgtcacatagtgaacgagc
If you have any comments or want to report a potential error in the tutorial, please email me (Chris Anderson) at JCAnderson2167-at-gmail.com