Arking:JCAOligoTutorial4
DBBS Method of Composite Part Assembly
The general idea of Biobricks is that you have a collection of basic parts, and then you assemble them into composite parts by some method of general assembly. DBBS (it stands for Digested Biobricks) is a PCR-based method that is very simple and efficient to implement. However, there are many other usable assembly methods. This or the three-antibiotic method are probably you're best choices for doing BBa-type assembly (XbaI/SpeI style). If you are doing BBb (BglII/BamHI), the 1-2-3 method is by far the best option, and will be described in a later tutorial section. The dbbs method assumes you have your basic parts in one of the following plasmids:
BBa BBb pSB1A2 pBca9145 pSB1A3 pBca1100 pSB1AK3 pBca1101 pSB1AC3 pBca1102 pSB1AT3 pBca1020 pBca1021 (and there will undoubtedly be more made)
All that is really necessary is that oligos ca998 and G00101 have annealing sites outside the Biobrick polylinker, and the Bla gene is present and in the same orientation in both plasmids.
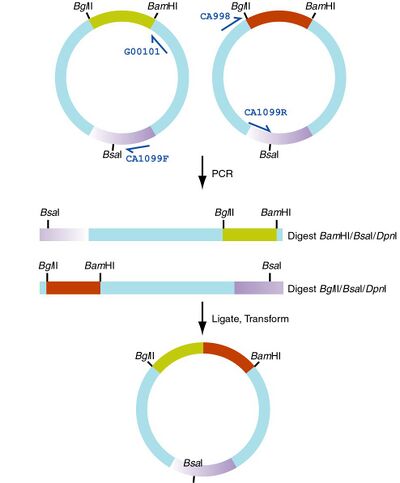
Generally, you're going to PCR amplify half the plasmid for the two parts you want to combine. You then digest the PCR products, ligate them together, and transform.
Implementation
Here is the general construction file for DBBS:
Oligo1 Oligo2 Template Digest ca1099F G00101 Left Part BamHI/BsaI/DpnI ca998 ca1099R Right Part BglII/BsaI/DpnI
Set up the two Expand PCRs on 25uL scale. Calculate the size of the product and run the appropriate program for that length (usually 4K55).
Run a Zymo column to remove the buffer and polymerase. Elute with 90uL water. Set up 100uL restriction digests as:
88uL DNA+water 10uL NEB Buffer 2 1 uL BamHI or BglII 1 uL BsaI 1 uL DpnI
Tap them to mix. It is imperative that you mix them well. If the DpnI doesn't get mixed, parent vector will bleed through resulting in parent vector side products. Incubate the reaction at 37deg for 1hr.
Run another Zymo column, elute the reaction in 30uL of water. These are the "dbbs".
Set up ligations:
6.5uL water 1uL 10x T4 DNA Ligase Buffer 1uL dbbs 1 1uL dbbs 2 0.5 uL T4 DNA Ligase
Transform, plate on a carbenicillin plate.
Storage
The dbbs digests will last forever if stored at -20. Since you only used 1 of 30uL of the material, you can do many assemblies with these parts. You'll log the dbbs into a google spreadsheet and place them in a special box for future use.
If you have any comments or want to report a potential error in the tutorial, please email me (Chris Anderson) at JCAnderson2167-at-gmail.com