IGEM:Caltech/2008/Project/Oxidative Burst: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
Doug Tischer (talk | contribs) |
Doug Tischer (talk | contribs) |
||
| Line 24: | Line 24: | ||
===Response=== | ===Response=== | ||
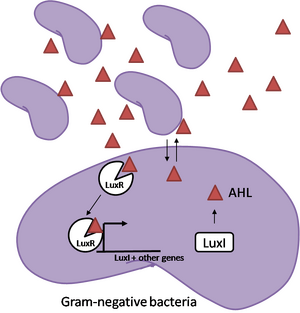
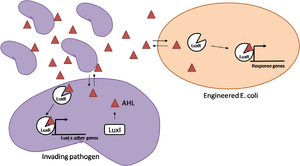
After sensing the presence of an invading pathogen, we want to engineer our E. coli to produce lethal amounts of hydrogen peroxide relatively quickly. We are not concerned with having the engineered E. coli survive either, as it is reasonable to assume that there are “unactivated” cells far away from the pathogen that could sustain the population. After LuxR binds AHL, it will activate transcription of an oxidase (an enzyme that produces hydrogen peroxide). There are many enzymes that can produce hydrogen peroxide in a stoichiometric ratio. The enzyme currently being used is galactose oxidase, as it has already undergone directed evolution to optimize its thermo stability and has been shown to be highly active in vitro. Other enzymes being considered are pyruvate oxidase and aldehyde oxidase. | After sensing the presence of an invading pathogen, we want to engineer our E. coli to produce lethal amounts of hydrogen peroxide relatively quickly. We are not concerned with having the engineered E. coli survive either, as it is reasonable to assume that there are “unactivated” cells far away from the pathogen that could sustain the population. After LuxR binds AHL, it will activate transcription of an oxidase (an enzyme that produces hydrogen peroxide). There are many enzymes that can produce hydrogen peroxide in a stoichiometric ratio. The enzyme currently being used is galactose oxidase, as it has already undergone directed evolution to optimize its thermo stability and has been shown to be highly active in vitro. Other enzymes being considered are pyruvate oxidase and aldehyde oxidase. | ||
Even though our engineered cells will eventually die from their oxidative burst, we want them to survive long enough to produce large amounts of the oxidase so they can produce large amounts of hydrogen peroxide. If the cells were left to produce hydrogen peroxide without any protection, they would produce just enough hydrogen peroxide to be cytotoxic and then fissle, killing only themselves but not much else. To avoid this problem, an E. coli catalase will be constitutively expressed, and then turned off shortly after the oxidase is being expreseed. We’re accomplishing this by putting katE (on of two E. coli catalase genes) behind the tetR sensitive promoter (tetR P) and having tetR co-transciptionally expressed with the oxidase. The time it takes for tetR to accumulate in the cell provides the delay in repressing katE expression. To ensure katE is rapidly cleared from the cells, it has a C-terminal ssrA degradation tag, which should reduce the protein’s half life to the order of minutes. In this way, the cell can be temporarily protected from hydrogen peroxide, but large amounts can accumulate before the substrate is exhausted. | |||
Even though our engineered cells will eventually die from their oxidative burst, we want them to survive long enough to produce large amounts of the oxidase so they can produce large amounts of hydrogen peroxide. If the cells were left to produce hydrogen peroxide without any protection, they would produce just enough hydrogen peroxide to be cytotoxic and then fissle, killing only themselves but not much else. To avoid this problem, an E. coli catalase will be constitutively expressed, and then turned off shortly after the oxidase is being expreseed. We’re accomplishing this by putting katE (on of two E. coli catalase genes) behind the tetR sensitive promoter (tetR P) and having tetR co-transciptionally expressed with the oxidase. The time it takes for tetR to accumulate in the cell provides the delay in repressing katE expression. To ensure katE is rapidly cleared from the cells, it has a C-terminal ssrA degradation tag, which should reduce the protein’s half life to the order of minutes. In this way, the cell can be temporarily protected from hydrogen peroxide, but large amounts can accumulate before the substrate is exhausted.The final strain will have deletions of both catalases, ensuring no interference. | |||
===System Design=== | ===System Design=== | ||
|} | |} | ||