3D DNA Patterning
Example Patterns
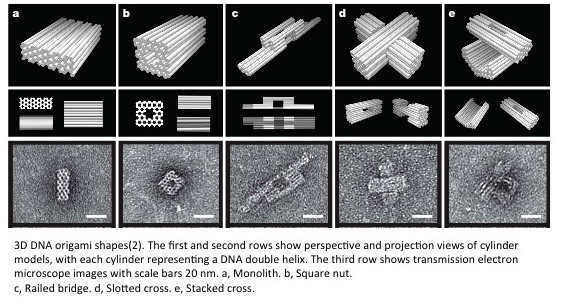
Specifications
| Theoretical | Empirical | |
|---|---|---|
| Pixel Dimension | 2 nm X 3.5 nm | 10 nm X 10 nm |
| Pattern Size | 300 nm X 300 nm | |
| Accuracy | 100% | 100% |
| Precision |
Mechanism
Deoxyribonucleic acids (DNA) are polymers made from monomers called nucleic acids. Nucleic acids are composed of the sugar deoxyribose with a 5’ triphosphate group and one of four nitrogenous bases (Adenine, Thymine, Guanine, or Cytosine). Two DNA strands can hybridize if their nitrogenous bases hydrogen bond. Guanine forms three hydrogen bonds with Cytosine and Adenine forms two hydrogen bonds with Thymine. The width of a double stranded helix in B-form is 2 nm. There are 10 to 10.5 nucleotides per helix turn with each nucleotide separated by ~3.4 angstroms.
DNA strands are assembled by slowly decreasing temperature until the DNA strands have found favorable energetic interactions. You can predict the critical temperature, called the melting temperature, at which half of the DNA molecules are hybridized by the amount of complementarity between two DNA strands. You do this by calculating the change in free energy of the reaction due to hydrogen bonding and entropy change of the system- although it is not that simple because cooperativity and volume exclusions effects play major roles in DNA assembly.
While the same DNA assembly method is used by everyone in the field, the architecture of the patterns differ and determine the pixel resolution, pattern size, precision, and accuracy. Precision and accuracy of the pattern are not measured in this field. Rather, the amount of defect free patterns over the total number of patterns is measured as yield. There are always many left over DNA strands that are washed away with this measurement. Here are some of the architectures I found:
- The first 3D DNA origami was a 22 nm diameter octahedron made with a 1.7 kilobase scaffold strand and five shorter strands(1).
- Shawn Douglas really extended 2D DNA origami to 3D DNA origami outside of polyhedra in 2009(2). Shawn’s design uses layers of helices in a honeycomb lattice in which staple strands crossover with holiday junctions achieving 200 nm shapes. However, the pixel size is around 20 nm X 20 nm and it takes a week to assemble. This architecture also allows bending the pixels up to 180 degrees and twisting the pixels up to 36 degrees(3). Ryosuke Iinuma was able to use the same strategy to create much larger patterns up to 300 nm in width(4).
- Another architecture is able to bend DNA into a full circle with a 40 nm diameter with 90% yields(5).
- Zhang et al. designed 3D wireframe shapes with 300 nm width and 10 nm pixel size while Benson et al. designed 3D wireframe shapes with 50 nm width and smaller pixel size(6, 7).
- 1 um X 80 nm DNA brick crystals can be created with pixels size 5 nm X 10 nm(8).
There are two major computational tools to design and simulate your DNA origami:
- caDNAno(9): This is an open-source design tool for 3D DNA origami using the honeycomb architecture.
- CanDO(10): This tool models 3D DNA structures based on DNA mechanical properties such as twisting, bending, and axial moduli, and entropic effects using statistical mechanics.
Contributions to understand yield optimization of 3D DNA origami:
- Sobczak et al. explored time and temperature effects of DNA assembly. They found that cooperativity effects allowed them to assemble DNA at a single temperature with higher yields(11).
- Wagenbauer et al. developed a tool to probe defects of origami. They designed 3 base pair probes that anneal to defect positions in the final shape. With the feedback from this too, yields of 99% were achieved(12).
- Chen et al. created a mechanical model for DNA origami. Mechanical effects of DNA elasticity play a major role in determining the right annealing temperature to assemble DNA(13).
I known these lists are not comprehensive. Please help me make these lists comprehensive.
Reference/Resources
- W. M. Shih, J. D. Quispe, G. F. Joyce, A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 427, 618–621 (2004).
- S. M. Douglas et al., Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 459, 414–418 (2009).
- H. Dietz, S. M. Douglas, W. M. Shih, Folding DNA into Twisted and Curved Nanoscale Shapes. Science. 325, 725–730 (2009).
- R. Iinuma et al., Polyhedra Self-Assembled from DNA Tripods and Characterized with 3D DNA-PAINT. Science. 344, 65–69 (2014).
- D. Han et al., DNA origami with complex curvatures in three-dimensional space. Science (2011), doi:10.1126/science.1201959.
- F. Zhang, Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nature Nanotechnology. 10, 779–784 (2015).
- E. Benson et al., DNA rendering of polyhedral meshes at the nanoscale. Nature. 523, 441–444 (2015).
- Y. Ke et al., DNA brick crystals with prescribed depths. Nature Chemistry, 1–9 (2014).
- S. M. Douglas et al., Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
- D. N. Kim, F. Kilchherr, H. Dietz, M. Bathe, Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 40, 2862–2868 (2012).
- J.-P. J. Sobczak, T. G. Martin, T. Gerling, H. Dietz, Rapid folding of DNA into nanoscale shapes at constant temperature. Science. 338, 1458–1461 (2012).
- K. F. Wagenbauer, C. H. Wachauf, H. Dietz, Quantifying quality in DNA self-assembly. Nature Communications. 5, 1–7 (2014).
- H. Chen et al., Understanding the Mechanical Properties of DNA Origami Tiles and Controlling the Kinetics of Their Folding and Unfolding Reconfiguration. J. Am. Chem. Soc. 136, 6995–7005 (2014).