2D DNA Patterning
Example Patterns
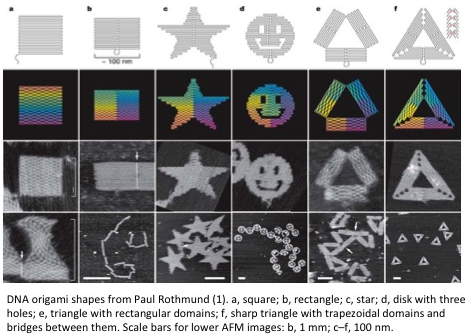
Specifications
| Theoretical | Empirical | |
|---|---|---|
| Pixel Dimension | 2 nm X 3.5 nm | 6 nm X 6 nm |
| Pattern Size | 100 nm X 100 nm | |
| Accuracy | 100% | 90% |
| Precision | 100% |
Mechanism
Deoxyribonucleic acids (DNA) are polymers made from monomers called nucleic acids. Nucleic acids are composed of the sugar deoxyribose with a 5’ triphosphate group and one of four nitrogenous bases (Adenine, Thymine, Guanine, or Cytosine). Two DNA strands can hybridize if their nitrogenous bases hydrogen bond. Guanine forms three hydrogen bonds with Cytosine and Adenine forms two hydrogen bonds with Thymine. The width of a double stranded helix in B-form is 2 nm. There are 10 to 10.5 nucleotides per helix turn with each nucleotide separated by ~3.4 angstroms.
DNA strands are assembled by slowly decreasing temperature until the DNA strands have found favorable energetic interactions. You can predict the critical temperature, called the melting temperature, at which half of the DNA molecules are hybridized by the amount of complementarity between two DNA strands. You do this by calculating the change in free energy of the reaction due to hydrogen bonding and entropy change of the system- although it is not that simple because cooperativity plays a major role in DNA assembly.
While the same DNA assembly method is used by everyone in the field, the architecture of the patterns differ and determine the pixel resolution, pattern size, precision, and accuracy. Paul Rothmund has achieved the best specifications for 2D DNA patterning(1). He is able to create patterns shown above by raster-filling the pattern with a long scaffold strand (7 kilobases) and by holding the scaffold together with shorter interweaving strands. While the precision and accuracy of patterning with Paul’s architecture isn’t great, other DNA assembly tricks can improve the accuracy and precision(2, 3). Another architecture is optimized for tiling 70 nm X 100 nm DNA rectangles into 10 um X 10 um lattices with 90% yield(4).
This field has been created by Nadrian Seeman and revolutionized in 2006 by Paul Rothmund(5).
Reference/Resources
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns. Nature. 440, 297–302 (2006).
- K. E. Dunn et al., Guiding the folding pathway of DNA origami. Nature. 525, 82–86 (2015).
- F. Dannenberg, K. E. Dunn, J. Bath, Modelling DNA Origami Self-Assembly at the Domain Level. arXiv (2015).
- S. Woo, P. W. K. Rothemund, Self-assembly of two-dimensional DNA origami lattices using cation-controlled surface diffusion. Nature Communications. 5, 4889 (2014).
- N. C. Seeman, An Overview of Structural DNA Nanotechnology. Mol Biotechnol. 37, 246–257 (2007).