20.109:TAP-tags
From OpenWetWare
Jump to navigationJump to search
New Protein Engineering Module for F08
Point of departure is to ask if tags are actually neutral wrt protein function
Rough sketch of labs
- Part 1: TAP tag
- non-essential SAGA subunits (essential likely in many complexes so purif profile complex + tag has many consequences)
- unknown ORFs changed during microarray of SAGA del or mtRNT1
- 1-1 Design primers
- 1-2 PCR
- 1-3 Transform
- 1-4 Confirm integration by colony PCR
- 1-1 Design primers
- Alternatively, could just give students tagged version.
- Part 2: Western
- need + TAP control
- use Sigma Ab to protA (anti-ProtA raised in rabbit or if need to detect tag when protA has been cleaved can use OpenBiosystems Ab)
- 1.5 ml ON culture, vortex 3X30' with glass beads and add 100 ul LB from Methods (2001) 24:218 barton link
- Part 3: Phenotypes
- compare parental, K/O, TAP-tagged
- examine GAL, Spt (his4-917d), TS, CS
- Part 4: Array
- compare parental to TAP-tagged
- Could also include
- Mass Spec analysis description/tour/lab?
- Tour of protein purif facility
- Mass Spec analysis description/tour/lab?
Experimental Details
Bacteria
- FB2305 = NB 311
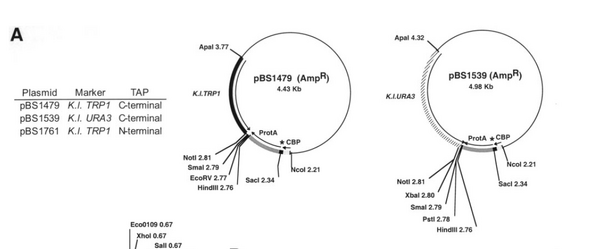
- MC1066+pBS1539 (AmpR+URA3-Kl, TAP tag)
- FB2306 = NB312
- NB318
- XL1-bl+pBS1479 (Lisa's prep of plasmid which does give PCR product for TAP-TRP, freshly transformed)
Yeast
- FY631 = Parental for TAP tagging = NY411
- MATa, his4-917d, lys2-173R2, leu2d1, ura3-52, trp1d63
- FY2714 = NY412
- MATa, HA-SPT3-TAP::TRP1, his4-917d, lys2-173R2, ura3D0 or -52, leu2D1 or D0, trp1D63
- FY2031 = NY413
- MATa, HA-SPT7-TAP::TRP1, his4-917d, lys2-173R2, ura3D0, leu2D1, trp1D63
- FY2305 = NY414
- MATa, SPT10-TAP::TRP1 trp1D63 his3D200 leu2D1 ura3-52 lys2-128d
- not his4-912d
- FY2306 = NY415
- MATa, SPT21-TAP::TRP1 trp1D63 his3D200 leu2D1 ura3-52 lys2-128d
- not his4-912d
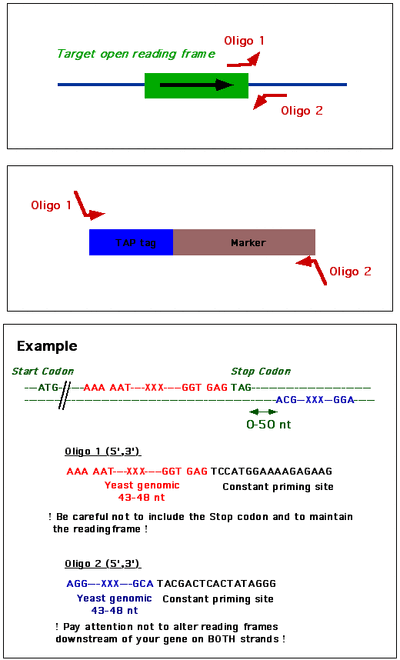
Primers for C-terminal fusions
General format for
- Upstream primer: 40nt then TCC ATG GAA AAG AGA AG
- Downstream primer: 40nt then TAC GAC TCA CTA TAG GG
Relevant sequence files
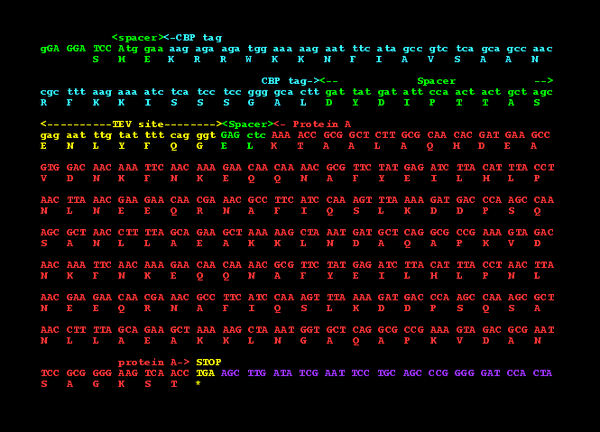
Needed primers
| needed to | forward primer (5' to 3') | reverse primer (5' to 3' |
|---|---|---|
| add TAP TRP to C-term of unknown 8 | NO222 GGT ATT TGT CGA TTA CAA AAC ACA TCC TGT AGG CGC AAA TTC CAT GGA AAA GAG AAG 57 mer Tm = 66.7 landing Tm (17bases only) = 45.5 |
NO223 GTT AGG ATT CAG TAC TAA CAC ATT CTC TAT GAC ACA ACC TTA CGA CTC ACT ATA GGG |
| check TAP | NO224 TCC ATG GAA AAG AGA AGA TGG AAA AAG AAT TTC ATA GCC GTC TC 44 mer Tm = 63.6 use with reverse primer from tagging PCR |
NO225 TCA GGT TGA CTT CCC CGC GGA ATT CGC GTC TAC 33 mer Tm = 67.9 use with fwd primer from tagging PCR expect length of TAP seq + TRP K. l seq = 554 bps (from 2147 to 2701 in pBS1479) |
| add TAP TRP to C-term of RNT1 | NO226 CTC ACA AAA GAA TAA GAA AAG AAA ATT CTC AGA TAC AAG CTC CAT GGA AAA GAG AAG 57 mer Tm = 63.8 |
NO227 ATC AAT GCA AGT TCC ATC ATG GTT GTG TAA AAG GAA CGT TTA CGA CTC ACT ATA GGG 57 mer Tm = 66.9 |
| longer landing sequences | TCC ATG GAA AAG AGA AGA TG = 20 mer fwd landing Tm = 49.6 ºC |
TAC GAC TCA CTA TAG GGC GA = 20 mer rev landing Tm = 54.3 ºC |
| Unknown 8 + longer landing sequences | NO228 GT ATT TGT CGA TTA CAA AAC ACA TCC TGT AGG CGC AAA TTC CAT GGA AAA GAG AAG ATG 59 mer Tm = 66.5 ºC |
NO229 TT AGG ATT CAG TAC TAA CAC ATT CTC TAT GAC ACA ACC TTA CGA CTC ACT ATA GGG CGA 59 mer Tm = 66.6 ºC |
| RNT1 + longer landing sequences | NO230 TCACAA AAGAATAAGA AAAGAAAATT CTCAGATACA AGC TCC ATG GAA AAG AGA AGA TG 59 mer Tm = 64.4 ºC |
NO231 TCA ATG CAA GTT CCA TCA TGG TTG TGT AAA AGG AAC GTT TAC GAC TCA CTA TAG GGC GA 59 mer Tm = 68.4 ºC |
| check TAP | NO258 for SPT3-TAP control TTT AAA GGT GGT AGA CTC AGT TCT AAA CCA ATT ATC ATG tcc atg gaa aag aga aga tg 59 mer use with NO225 = reverse primer for checking TAP tag expect length of TAP seq + TRP K. l seq = 554 bps (from 2147 to 2701 in pBS1479) |
NO259for SPT7-TAP control AAT AGT TCA TTT AGC TTG AGC CTT CCT CGC CTT AAT CAA tcc atg gaa aag aga aga tg 59 mer use with NO225 = reverse primer for checking TAP tag expect length of TAP seq + TRP K. l seq = 554 bps (from 2147 to 2701 in pBS1479) |
| Check TAP with NO224 | SPT3_check_rev NO268 ACC AGA AGG AAA CCC ATG CAC CTC CAT GAT GAA ATT ATA CAA AAA T |
SPT7_check_rev NO269 AGC AAA TGT ATT TAA TTA AAG ATA AAA TAT TCA ACT ATT TAG CGC GCT CA |
Genes to tag
Option 1: S. cerevisiae SAGA subunits
| Subunit | size,chromosome,null p-type |
|---|---|
| Ada subunits | |
| Ada1 (aka HFI1, SUP110, SRM12, GAN1) | 1.467 kb=489 aa, Chr. XVI, viable |
| Ada2 (aka SWI8) | 1.305 kb=434aa, Chr. IV, viable |
| Ada3(aka NGG1, SWI7) | 2.109 kb=702aa, Chr. IV, viable |
| Gcn5 (aka ADA4, SWI9) | 1.32 kb=439aa, Chr. VII, viable |
| Ada5 (aka SPT20) | 1.815 kb=604aa, Chr. XV, viable |
| Spt subunits | |
| Spt3 | 1.014 kb=337aa, Chr. IV, viable |
| Spt7(aka GIT2) | 3.999 kb=1332aa, Chr. II, viable |
| Spt8 | 1.809 kb=602aa, Chr. XII, viable |
| Spt20 (aka Ada5) | 1.815 kb=604aa, Chr. XV, viable |
| TAF subunits | |
| TAF5 (aka TAF90) | 2.397 kb=798aa, Chr. II, inviable |
| TAF6 (aka TAF60) | 1.551 kb=516aa, Chr. VII, inviable |
| TAF9 (aka TAF17) | 0.474 kb=157aa, Chr. XIII, inviable |
| TAF10 (aka TAF23, TAF25) | 0.621 kb=206aa, Chr. IV, inviable |
| TAF12(aka TAF61, TAF68) | 1.620 kb=539aa, Chr. IV, inviable |
| Tra subunit | |
| Tra1 | 11.235 kb=3744aa, Chr. VIII, inviable |
| Other subunits | |
| Sgf73 | 1.974 kb=657aa, Chr. VII , viable |
| Sgf29 | 0.779 kb=259aa, Chr. III, viable |
| Sgf11 | 0.3 kb=99aa, Chr.XVI, viable |
| Ubp8 | 1.416 kb=471aa, Chr. XIII, viable |
| Sus1 | gene with intron, Chr. II, viable |
Option 2: Unknown ORFs from SGF73/sgf73 array
| Unknown | ORF | SGF73 green signal | sgf73 red signal | log2 (green/red) |
|---|---|---|---|---|
| 1 | YHR033W | 38938 and 69586 | 285 and 570 | 7.1 and 6.9 |
| 2 | YOR302W | 3374 and 6054 | 49 and 167 | 6.1 and 5.2 |
| 3 | YJR097W | 524 and 1052 | 13 and 28 | 5.3 and 5.2 |
| 4 | YBL028C | 1146 and 2706 | 32 and 323 | 5.2 and 3.1 |
| 5 | YDR034W-B | 17290 | 447 | 5.3 |
| 6 | YGR067C | 12025 | 320 | 5.2 |
| 7 | YKL037W | 8340 | 282 | 4.9 |
| 8 | YER067W | 6296 and 12450 | 82556 and 81036 | -3.7 and -2.7 |