Yi-An, Spring 2006
Friday, March 3, 2006
Perry and I transformed the DH5alpha competent cells with pCMV-Tag2a vectors. DH5alpha cells were used because they have higher efficiency in transformation. We placed 50 uL of DH5 alpha cells with 1 uL of the pCMV-Tag2A vectors (which was a 1 uL/uL stock) into a 1.5 mL eppendorf tube. This was repeated for a second transformation (2 pCMV parallel transformations were performed). For the positive control, 2.5 uL of pUC19 was placed into a 1.5 mL eppendorf tube along with 50 uL of DH5alpha competent cells. These cells were placed on ice for 30 minutes. Subsequently, we heat shocked the cells at 42 degrees Celsius for 25 seconds then placed them on ice for 2 additional minutes. SOC medium was incubated and 950 uL were added to each eppendorf tube. The tubes were then placed into the shaker (225 rpm, 37 degrees celsius) for an hour. The protocol used modeled that obtained from the DH5 product information sheet).
We then transformed TOP10 competent cells with hD1g-cloned Topo vectors: SG25, SG35, and F11. We diluted the stock vector 1:10. 5 uL of each of the necessary vectors were added to separate tubes of TOP10 competent cells. 2.5 uL pUC19 was added to one tube of top10 cells to serve as the positive control. Nothing was added to the tube of competent cells - this served as the negative control. The tubes were set on ice for 30 minutes, heat shocked for 25 seconds, set on ice for another 2 minutes, and then placed in the shaker for an hour (225 rpm at 37 degrees Celsius). Perry plated the transformations.
Protocol used for the second step was one i wrote up from mcb100 (included below):
1) Grab an ice bucket and fill it with ice.
2) Obtain 2 tubes from the "One Shot Top 10: Chemically Competent Cells" box found in the bottom shelf of the upper -80 Degree Celsius Freezer.
Obtain: - 1 purple tube - cells
- 1 black capped tube - plasmid (for positive control)
3) Label 3 eppendorf tubes and initial:
(+) for positive control with plasmid (-) for negative control with water LIG for ligation DNA (you can use the whole cell vial for this)
4) Pipet 10 - 15 microliters of cells from the purple capped vial into each eppendorf tube.
5) Place 2 microliters of positive (plasmid), (-) water, and ligation DNA into their respective labeled tubes
6) Place the 3 tubes in the ice bucket and set a timer for 15 minutes
7) Place the tubes in the heat plate at 42 degrees Celsius for 30 seconds (Set a timer).
8) Place the tubes back in the ice bucket for 2 minutes (set timer).
9) Pipet 250 microliters of S.O.C. Medium (yellow fluid) into each eppendorf tube.
10) Place the 3 eppendorph tubes in the incubator in the other room (near the computer - to the left) for 45 - 50 minutes.
11) Obtain 3 Amp resistant petri plates (found in the back refrigerator of the other lab room). Label the plates with (+), (-), and LIG.
12) Obtain the 3 incubated vials from the other room. Get glass beads from the back shelf by the window.
13) Pipet the fluid from each vial into their respective petri plates (pipet onto the center of the plate).
14) Pour a few (approximately 10 beads) onto each plate. Cap the plates and shake them side to side [avoid shaking them in a circular manner because this will cause a higher concentration of bacteria to end up in the center of the plate. You want them to be spread out as evenly as possible). Place the used beads in a small beaker, rinse, and place them in the dirty equipment tub for cleaning.
15) Cover the plates and place them in the incubator overnight.
Saturday, March 4, 2006
I came into biolabs tonight and performed another transformation on the DH5alpha competent cells with pCMV-Tag2a vectors as well as another transformation of the TOPO vector (F11, SG25, SG35) with TOP10 competent cells. The same protocol (above) was used for the respective transformations.
Sunday, March 5, 2006
Perry performed an innoculation using the successful transformations from one of the previous transformations.
Tuesday, March 7, 2006
The innoculation yielded very little pellet, thus, we made a Starter culture for pCMV and SG25. 3 mL of LB media was used for each labeled Falcon tube. We placed 1.8 uL of KAN in the pCMV tube and 1.0 uL of Amp in the SG25 tube. A sterile loop was used (note: yellow rods, found in the other room, can be used in place of the sterile loops) to pick a colony from the pCMV transformation plate. The sterile loop end was swooshed in the tube. 10 mL of the SG25 glycerol was added to the respective eppendorf tube. Both tubes were placed in the 37 degree celsius, 225 rpm shaker. The QIAGEN Plasmid Purification Handbook (p.20) states that the incubation period should last 8 hours.
Note to self: for the innoculation, use the LB medium located on the back (far right) shelf (the terrific broth was contaminated).
Protocol for Innoculation: Dilute the starter culture - 200 mL LB, 400 uL culture, 120 uL (KAN - for pCMV) or 200 uL (Amp = for SG25). Grow at 37 degrees Celsius for 12 - 16 hours in the 225 rpm shaker.
Wednesday, March 8, 2006
The innoculations yielded substantial pellets. Alain spun down the pellets and I performed the midiprep.
Midiprep Protocol notes (for future reference):
1) centrifuge the innoculation 6000 rpm for 15 min at 4 degrees Celsius
2) resuspend the bacterial pellet in 4 mL Buffer P1 (use the vortex machine to break up the pellet. Then transfer the pellet to a smaller centrifugation tube (the one we used in mcb100 - clear/transparent tubes)
3) Add 4 mL Buffer P2, mix gently through inverting the tubes 4 - 6X. Incubate at rm temperature for 5 minutes
4) Add 4 mL of chilled P3 buffer, mix gently (invert 4 - 6 X), incubate on ice for 15 minutes.
5) Centrifuge at 13,000 rpm at 30 minutes at 4 degrees Celsius. Remove supernatant. (note to self: mark the edge of the tube which will be facing up in the centrifuge - just so that you can locate the pellet with greater ease later in the procedure).
6) In the meantime, equilibrate the QIAGEN-tip 100 (found in the Midiprep kit) by applying 4 mL of QBT buffer to each column. (i used 4, there is also a new rack - transparent blue - that nullifies the need to set up separate racks - found in room 5088)
7) Apply the supernatant from the centrifugation step to the QIAGEN-tip. After it has flowed through, wash the QIAGEN-tip with 2 X 10 mL QC buffer (also found in the midiprep kit)
8) Get a new set of centrifugation tubes - the clear, opaque ones. Elute the DNA into these tubes with 5 mL QF Buffer.
9) Precipitate the DNA by adding 3.5 mL room temp. isopropanol to the eluted DNA. Mix and centrifuge at 11,000 rpm for 30 minutes at 4 degrees Celsius. Decant the supernatant.
10) Wash the DNA pellet with 2 mL of room temp 70% ethanol (do not disturb pellet). Decant the supernatant (do not disturb the pellet).
11) Air dry the pellet for 5 - 10 minutes. Redissolve the DNA in 250 - 300 uL of distilled H2O.
Performed nanodrops for each of the construct midipreps. Two samples were measured for each midiprep. The results are as follows:
F11: 604.9 ng/uL and 615.2 ng/uL SG25: 229.3 ng/uL and 228.9 ng/uL SG35: 89 ng/uL and 90.4 ng/uL pCMV: 213.5 ng/uL and 212.4 ng/uL
- all the measurements displayed the appropriate 260 nm peak.
Also, Alain showed me how to use the new PCR machine located in room 5096. We programmed "BB2" which stands for BioBricks 2, a simplification due to the inefficiencies of the machine in typing in letters.
The program consists of the following steps:
Step 1: 94 degrees, for 5 minutes
Step 2: 94 degrees, for 30 seconds
Step 3: 50 degrees, for 30 seconds
Step 4: 68 degrees, for 1 minute
Step 5: repeat steps 2 through 5 for 30 cycles
Step 6: hold at 4 degrees, forever (typed in 0 to allow for this)
Step 7: END
Functions of the machine:
- the buttons (<-) and (->) allow you to scroll through options, or input a "space"
- the button (^) allows you to select something that you want
Prepared 4 samples of PCR reactions with the following composition: general formulation: 45 uL PCR supermix, 2 uL distilled water, 1 uL of primer1, 1 uL of primer2, and 1 uL of diluted template.
- Part1: F11 (template) and primers (L27S + L27A)
- Part 2: SG25 (template) and primers (SH3S + GKA)
- Part 3: SG35 (template) and primers (SH3S + GKA)
- Part 4: SG35 (template) and primers (SH3S + I3A)
I diluted the SG25 and F11 in a 1:100 proportion and performed a 1:10 dilution of SG35 (as a result of the aforementioned concentrations obtained for the different constructs.
- Note: All dilutions and templates are located in the large -20 freezer in room 5096 in the clear box on the right side of the second shelf. All primers are located in the green labeled box on the right side of the 2nd shelf in room 5096.
Thursday, March 9, 2006
Retrieved the PCR from last night and ran a mini-gel (2% gel) with the 4 PCR samples and 2 ladders (the 1Kb ladder and the 2 log ladder - both of which can be found in the normal mcb100 project 3 freezer in room 5088)
Results:
For the minigel run:
- make sure that all volumes in all wells equals 20 uL. 10 uL of PCR DNA was used for each sample and 10 uL of each ladder was used. 10 uL of water was added in a separate eppendorf tube to mix either ladder or DNA with water. Water was added to all empty wells (20 uL)
- explanation of each of the gel usages (to come): 0.8%, 1.2%, v. 2%
Performed a PCR Purification (protocol):
1. Add 5 volumes of Buffer PB to 1 volume of PCR sample and mix. (200 uL of PB was added)
2. Apply the sample to a QIAquick spin column and centrifuge at 13000 rpm for 1 min. discard flow-through.
3. Replace the column. Add 750 uL PE Buffer and centrifuge for 1 minute. Discard flow through. Replace the column and centrifude for another 1 minute
4. place each column in a clean/new 1.5 mL eppendorf tube. Elute with 30 uL dH2O, let the column stand for 1 minute, centrifuge for 1 minute.
Performed TOPO Cloning:
1) Get the necessary number of tubes (in this case 4 tubes) of Chemically Competent Top10 Cells, place on ice (for use in transformation).
2) get 4 eppendorf tubes (label appropriately #1-4)
3) Create the following mixture: 4 uL Fresh PCR product, 1 uL Salt solution, 1 uL of TOPO vector.
4) Incubate the tubes at room temperature for 5 minutes
5) set on ice then procede to transformation
For the transformation: i followed the proctocol above (taken from my old mcb100 wiki page)
- Diluted the I2-I5-I4 TOPO (1:100)
Performed PCR reaction for the last 5 parts using program BB2:
- Part 5: SG25 (template) and primers (I2S + GKA)
- Part 6: F11 (template) and primers (SH3S + SH3A)
- Part 7: I2-I5-I4/Topo (template) and primers (I2S + I4A)
- Part 8: F11 (template) and primers (GKS + GKA)
- Part 9: I2-I5-I4/Topo (template) and primers (I5S + I4A)
Friday, March 10, 2006
- the transformations that I performed last night did not yield any colonies. There seemed to be a weird bumpy lawn of (something) on the agar; however, they were not colonies.
- Thus, i performed ran an e-gel of the purified PCR DNA (using 10 uL dna and 10 uL water for each well sample). The e-gel showed that there were DNA bands in all expected lanes for the roughly the expected band lengths. The bands however, seemed a bit light/dull.
- Perry performed the cloning, transformation, and purification of parts 5 through 9. Two parts were successful, he re-PCR'd the other 3 parts.
Saturday, March 11, 2006
- Performed another TOPO cloning procedure on Parts 1 through 4. I included a positive control (using pUC 19 cells that were located in the -80 freezer) as well as a negative control (with nuclease free water).
- Performed a transformation on the 6 samples.
Monday, March 13, 2006
- We performed a transformation of SG35 with different concentrations/volumes. We prepared a positive and negative control using the TOP10 competent cells (2 tubes). Instead of adding 250 uL of SOC medium, we added 350 uL to each tube, which we plated onto 6 CARB plates. For both positive and negative controls, we plated the transformation in the following amounts: 50 uL, 100 uL, and 200 uL.
- We left the plates in the incubator overnight.
- Perry performed a digest of the pCMV vector.
Tuesday, March 14, 2006
- We came into lab today to do a few things.
- Perry ran a gel of the pCMV digest and gel extracted the DNA.
- We discovered that our transformations did not work - the strange bacterial lawn growths on all of the plates (both positive and negative) indicated that the CARB amount in the agar was not strong enough. The lack of selection indicated that new plates had to be made (Alain did this).
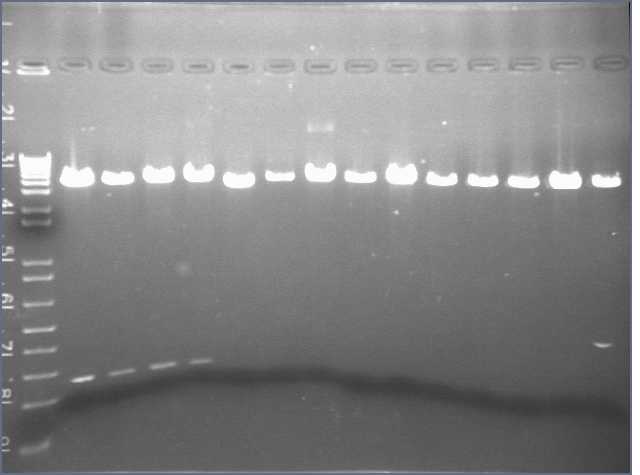
- I performed another PCR reaction for each of the 9 parts. After the 30 cycles, i used a gel Alain prepared and ran a gel of the samples (20 sample well comb). I added 5 uL of loading dye to each PCR sample.
The well contents from left to right:
- Lane1: 1 Kb ladder (10 uL)
- Lane 2 & 3: Part 1(23 uL each)
- Lane 4 & 5: Part 2 (23 uL each)
- Lane 6 & 7: Part 3
- Lane 8 & 9: Part 4
- Lane 10 & 11: Part 5
- Lane 12 & 13: Part 6
- Lane 14 & 15: Part 7
- Lane 16 & 17: Part 8
- Lane 18 & 19: Part 9
- Lane 20: 2 log ladder
Picture to come :)
- I excised the gel bands from the gel. Perry and I performed a gel extraction, DNA purification. topocloning and transformation.
Wednesday, March 15, 2006
- We performed a transformation of pCMV Tag2 control and pCMV Tag2a. We also included a positive and negative control.
- I performed an innoculation of Parts 1 through 9 using colonies from last night's transformation. I used the plates that Alain made with the Ampicilin antibiotic (located in the last fridge in room 5096).
Thursday, March 16, 2006
Perry performed the minipreps for parts 1 through 9. He also performed the respective digests.
Friday, March 17, 2006
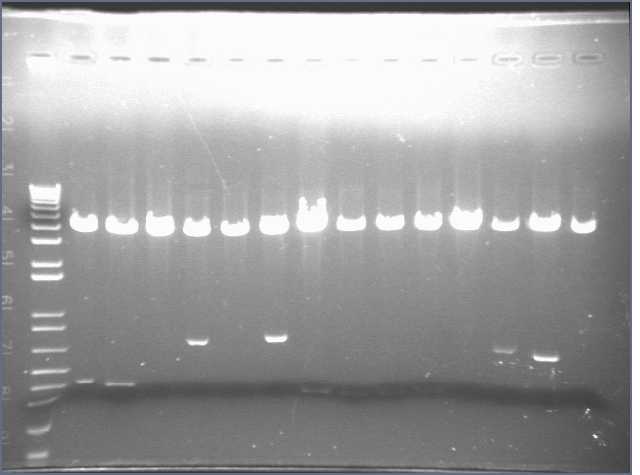
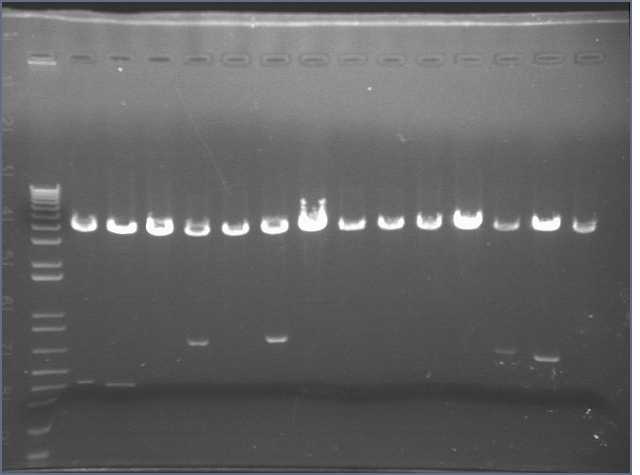
We came in this morning and ran gels of the 28 digests. The gel run yielded bands for parts 1, 4, 5, 6, and 8. Parts 2 and 3 did not display bands. Part 6 exhibited the wrong band length.
As a result, I performed a PCR reaction for parts 2, 3, and 5. I ran a gel of the PCR products, however this did not yield any results. Part 3 showed up faintly on the gel. Bands were absent for Parts 2 and 5.
Alain and I innoculated the pCMV-Tag2a, pCMV Tag2, Parts 2, 3, and 5. We innoculated 4 tubes of each of the parts and 2 tubes of pCMV Tag2a and pCMV Tage 2. For parts 2, 3, and 5 i innoculated 3 mL of LB per tube with 3 uL of Amp in each tube. I streaked the selected bacteria on an Amp plate. The plate was placed in the incubator and the 16 tubes were placed in the shaker.
I discovered via email and re-examination of the pcr pictures that i was not supposed to pcr part 5 or innoculate part 5 for that matter.
Thus, as a result, i innoculated part 6. I chose 4 colonies, streaked, and innoculated them.
I reperformed the PCR reactions for Parts 2, 3, and 6 using program BB3. Let the reaction run overnight.
Saturday, March 18, 2006
- Ran a gel of the three PCR parts - parts 2, 3, and 6. Only part six displayed visible bands. *picture to come*
- performed minipreps on the following parts: P2-5, P2-6, P2-7, P2-8, P3-5, P3-6, P3-7, P3-8, P5-5, P5-6, P5-7, P5-8, P6-5, P6-6, P6-7, P6-8
- performed restriction enzyme digests on each of the miniprepped parts. Used 10 uL of each miniprepped dna. Made a mastermix of 18 uL EcoRI, 45 uL 10x EcoRI buffer, 4.5 uL BSA, and 202.5 uL H2O. 15 uL was alliquoted to each digest tube.
(for single digest reactions: use 1 uL EcoRI, 2.5 uL 10X EcoRI buffer, 0.5 uL BSA, and 11.25 uL H2O)
- Alain recommended that since parts 2 and 3 did not yield visible gel bands that it may be a problem with the transformation. Possibly that the Top10 Competent cells do not work well with pCMV, thus tomorrow i will try to transform the pCMV Tag2 and Tag 2A again using DH5alpha.
- the pCMV innoculations did not really grow, or they are growing they are growing at a snail's pace.
Tuesday, March 21, 2006
- performed a miniprep on spun down pellets of our overnight innoculations for pCMV Tag2 and pCMV Tag2a.
- continued to perform a restriction enzyme digest with BamHI and Xho1. I used 5 uL of the DNA, 2 uL of BamHI buffer, 0.5 uL BamHI, 0.5 uL of Xho1, and 12 uL of water. The samples were placed in the PCR machine and run under the program "37 degrees - 6 hours"
- Alain ran a PCR gel and turned up fabulously bright bands :) He took those and excised the appropriate bands.
- I took the the gel isolated bands and performed a QIAEX gel extraction. The incubated tubes were pink/orange in color for an extended period of time - i added alliquots of 10 uL sodium acetate to attain a yellow pigment fluid (pH = 7.5).
- Subsequently, I topo-cloned the DNA samples (4 uL dna, 1 uL salt solution, 1 uL vector) and performed a transformation (which i left in the 37 degree incubator overnight).
Wednesday, March 22, 2006
- Alain ran a gel of the digests (5 uL of digest was used in each well) that i had placed in overnight. The gel yielded the following results:
- picture to come - my email isn't working properly*
- the transformations from the previous night for P2, 3, 5 yielded many colonies. Thus, i performed innoculations of 4 colonies per part. I streaked the colonies onto a master plate and placed the plate in teh 37 degree incubator. I placed the innoculations in the shaker overnight. The innoculations were 2 mL LB, 2 uL Amp, and a toothpick swab of a colony.
- performed another restriction digest of pCMV Tag 2 and 2a due to successful gel results. For these digests I used, 20 uL dna, 5 uL BamHI Buffer, 1 uL BSA, 1 uL Xho1, 1 uL BamHI, and 22 uL of Nuclease Free water (for 50 uL digest reactions).
Thursday, March 23, 2006
- performed a miniprep for Part 5 (parts 2 and 3 were discarded due to the discovery that 2 primers had mutations in them - GKA and LH5)
- will return later to complete the gel extraction, topo cloning, and transformation for pCMB?V Tag 2a.
Thursday, April 6, 2006
Back from spring break!
Today, perry and i performed a midiprep of P1-1a and P2-3a.
- performed 2 sets of innoculations. The first set of 4 consisted of streaking and innoculating colonies from a transformation that perry performed yesterday (P1-1a). The second set of innoculations consisted of Part 3 Topo II (4 clones were selected) and pCMV Tag2 BB linker (4 clones were selected).
- prepared sequencing reactions for Parts 5, 7, 8, 9 (specifically Part 5-1a through 4a, Part 7 - 2a, Part 8- 1a through 3a, Part 9 - 1a, 3a, and 4a). The tubes were labeled 1 through 11 respectively (in the order mentioned previously). 8 uL of DNA were placed in each sequencing tube.
Wednesday, April 12, 2006
Performed an innoculation for BB3 (from the streaked plate). Sequences BB3 and BB4 were both accurate.
Next step: - run gels of the digests (mini and midipreps digests); the digests for the midipreps should be gel excised, miniprep digests can be run through an egel. - midiprep of BB3
Thursday, April 13, 2006
- the innoculation was not successful. Thus, i prepared 2 3mL cultures of BB3 and BB4 (using KAN - 2 uL).
- spun down the innoculations of p3-12a, P7-2a, P9-3a. Will midiprep all of these parts and the bblinker upon successful innoculation of that last part.
- perry ran two gels of the digests performed yesterday. We took pictures of the results and excised the bands.
- performed a gel extraction of the isolated gel pieces - stored in my box.
- performed a transformation using DHalpha cells for the BB3 linker. Performed a positive and negative transformation alongside that of the BB3 linker.
Friday, April 14, 2006
- performed a 200 mL culture of BB3 (both BB3 and BB4 3mL innoculations were successful) using 200 mL LB, 120 uL (written on the KAN bottle - 60 uL/100mL), and the innoculation for BB3 from last night.
- performed another 2 mL falcon tube innoculation of the BB3 linker (transformation with DH5alpha)
- last night's tranformations were all successful - yielded two plates full of colonies.
Saturday, April 15, 2006
- made a glycerol stock of the the BB3 linker (with DH5alpha). Took the 2 mL innoculation and split it into two 1.5 mL tubes (1.0 mL of innoculation per tube), spun these down, and added 400 uL of 20% glycerol/LB to each tube
- performed a midiprep of the 4 parts (the BB3 innoculation was successful) - BB3, P3-12a, P7-2a, and P9-3a
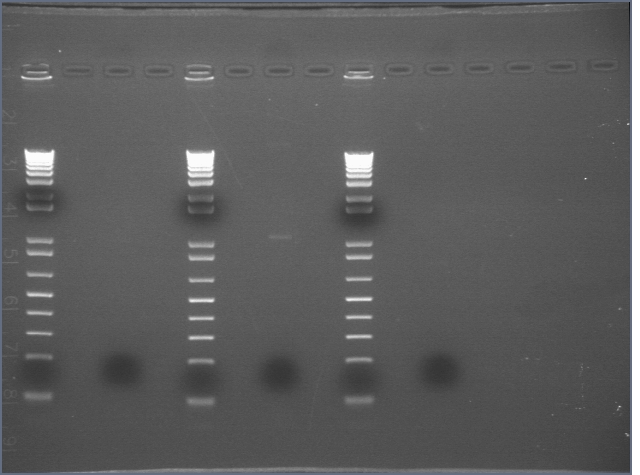
- made 1:10 (50 uL total) dilutions of the midi's in order to nanodrop
- nanodropped the dilutions:
- P9-3a: 57.3 ng/uL (peak at 260 nm)
- P7-2a: 25.3 ng/uL (peak at 260 nm)
- P3-12a: 66.1 ng/uL (peak at 260 nm)
- BB3: 77.8 ng/uL (peak at 260 nm)
- performed a restriction enzyme digest (placed it in pcr machine 5). for parts 3, 7, and 9 i used:10ul DNA, 5ul buffer3, 0.5ul BSA, 1.25ul XbaI, 1.25ul PstI, 32ul water.
for the BB3 linker i used: 10ul DNA, 5ul buffer2, 0.5ul BSA, 1.25ul SpeI, 1.25ul PstI, 32ul water.
Monday, April 17, 2006
- we ran the digests from saturday. Some of the bands blurred into adjacent bands. - Perry performed a second digest. The gel pictures indicated that i had used too much DNA (used 10 uL of the midi when i probably should have used the 1:10 dilution of the midi).
Tuesday, April 18, 2006
- made and ran a gel of all the digests from yesterday. - gel isolated the fragments - performed a gel extraction
- placed the gel extractions/digests in a box "PT/YK"
- (1) P1-1a
- (2) P2-3a
- (3) P3-12a
- (4) P4-4
- (5) P5-4a
- (6) P6-4
- (7) P7-2a
- (8) P8-2a
- (9) P9-3a
- P1 (delta E1/S1)
Wednesday, April 19, 2006
- performed a restriction digest (PstI on the BB3 part that had been singly digested with SpeI) - prepared a double digest with EcoRI and SpeI (used EcoRI buffer); both digests were set at 37 degrees celsius for 4 hours -treated each of the tubes of BB3 with CIP (placed it in the PCR machine for 1 hour at 37 degrees - program DIGEST) - made and ran a gel of the BB3 samples (picture to come) -gel isolated the pieces
Friday, April 21, 2006
- performed a gel extraction of all BB3 gel samples (4 gels total) - performed a ligation of P1 and BB3; stored in the refrigerator - attempted to make a gel but took out the comb too early so the gel wells collapsed... making and running the P1 (delta E1/S1) digest gel tomorrow.
Saturday, April 22, 2006
- made and ran a gel for P1 digest (delta E1/S1); no results - the gel was blank... - reperformed digests for P1 with the following enzyme pairings: Xba1/PstI or EcoRI/SpeI.
Tuesday, April 25, 2006
- made and ran a gel of the digests P1-1a with XbaI/PstI and EcoRI/SpeI
- gel isolated the pieces
- performed gel extraction of the pieces; froze the samples
- the bands on from the aformentioned gel isolation were not very bright; reperformed the P1-1a digest (2 tubes: 1 tube with restriction enzymes XbaI/PstI, the other tube with enzymes EcoRI/SpeI). These new digests included 30 uL of 1:10 diluted P1-1a midiprep DNA and 12 uL of water. 0.5 uL BSA, 1.25 of each restriction enzyme, and appropriate buffer was used (buffer 3 and EcoRI buffer respectively).
- performed a transformation of the ligation that i performed last friday. Used DH5alpha cells and only added 400 uL of SOC media. All of these transformations were plated on KAN plates. No positive control was made.
Wednesday, April 26, 2006
- the transformation from yesterday yielded no colonies.
- made and ran a gel of the digests of P1-1a (4 tubes total: with XbaI/PstI or EcoRI/SpeI)
- gel isolated the pieces
- performed a QIAEXII gel extraction
- performed two ligations: one ligation (ligation #1) was with P1-1a (digested with XbaI/PstI) and pCMV-tag2BB (which was digested with SpeI/PstI); the other ligation (ligation #2) was with P1-1a (digested with EcoRI/SpeI) and P3 (which was digested with XbaI/PstI). Since I didn't know what ratio of insert:vector to use, Alain suggested the following: one ligation with 5 uL P1 and 5 uL of P3, and another ligation with 4 uL of P1 and 1 uL of P3.
- performed a transformation on Ligation #1 and plated the transformation on KAN plates.
- set up a PCR reaction for Ligation #2. To test the optimal amount of DNA to place in each reaction tube, i set up a total of 8 reaction tubes (2 sets of 4 tubes with varying amounts of ligation DNA included - 1 uL, 2 uL, 4 uL, or 6 uL)
Thursday, April 27, 2006
- the transformation from yesterday was unsuccessful.
- As a result, I ran the digested P1 (with XbaI/PstI), P3 (with XbaI/PstI digest), and BB3 (with SpeI/PstI digestion) along with the PCR reactions from yesterday. (picture to come). The gel showed that P1 was either very light (barely visible) or not there.
- Consequently, I re-performed the P1-1a digest with XbaI/PstI (since there was no more left) - two digests were performed.
- Alongside this, i also performed a digest of the P1-P3 construct with XbaI/PstI.
- To start on the construction of the other parts, I ligated P1 to P2, P1 to P4, and P1 to P6.
- I set up 2 sets of PCR tubes for each ligation (6 PCR reaction tubes total). For the PCR reaction, one tube with 1 uL ligation DNA and another with 4 uL ligation DNA was tested. (5 uL of the each of the appropriate primers were utilized). For the P1-P2 PCR reaction, I used L27S and GKA. For the P1-P4 PCR reaction, I used L27S and I3A. For the P1-P6 reaction, I utilized L27S and SH3A.
Friday, April 28, 2006
- ran an e-gel of the PCR reactions from last night (used 10 uL of DNA and 5 uL of 50% glycerol + TBE) for P1-P2, P1-P4, and P1-P6. The gel yielded clear bands for each of the PCR constructs.
- performed a PCR purification of the PCR samples that i didn't run through the e-gel using the QIAquick PCR purification set
- performed a restriction enzyme digest with XbaI/PstI for each of the three constructs using the DIGEST program on PCR machine 3
- made and ran a gel for the digests performed last night {P1 (delta X1/P1) and P1-P3 (delta X1/P1)}
- gel isolated the pieces and performed a gel extraction using the QIAEXII protocol
- performed a ligation of the P1-P3 part with the pCMV-tag2BB (BB3) which was previiously digested with SpeI and PstI. Since i didn't know the proper ratio for the the insert:vector. I performed two separate ligations based on the strength of the bands on the gels for each construct. The P1-P3 construct displayed strong bands while the BB3 displayed weaker bands. One ligation was performed with 5uL P1-P3 (delta X1/P1) & 5 uL BB3 (delta S1/P1) while the other ligation was performed with 3 uL of p1-P3 and 7 uL BB3.
- i diluted 1 uL of the pCMV-tag2BB midiprep 1:100.
- trasnformed 2 uL of the diluted BB3midiprep along with the other two ligations. The transformation of the midiprep dilution serves as a positive control (since it contains the proper resistance cassette). I used KAN LB Min plates since we were out of regular KAN plates.
Monday, May 1, 2006
Tuesday, May 2, 2006
- made and ran a gel of the overnight digests - gel isolated and extracted the pieces - ligated the following together:
- P1-P2 to pCMV
- P1-P3 to pCMV-tag2BB