User talk:Joseph P. Torella
Summary of My Contributions to the Class Project // 12-08-09::12-17-09
I'll be updating this section somewhat as the semester wraps up (as Harris has said this should be done by the end of semester, rather than by this Thursday). I'll try to put each of my contributions below, plus what I think are the most important lessons learned, or ideas if the project were to continue:
- I proposed the use of Logistic Models to predict disease risk as a function of multiple SNPs, which ultimately became important in the modeling group's attempts to rationally produce such models from GWAS data.
- I produced a small command-line utility, TraitFinder, to parse a single genome on trait-o-matic for all SNPs belonging to a particular disease/trait, extract their rsid's and odds ratios, and print them out. Although this has not been as generally useful as the infrastructure group's great work on interfacing directly with trait-o-matic on the web, the tool may still be useful for those who want to do their own analyses.
- The logistic model idea came from literature review I did on Multi-SNP Predictions of Disease Risk, especially on Prostate Cancer, where family history and five SNPs together account for 50% of overall Population Attributable Risk (PAR) of prostate cancer.
- Since I have some familiarity with both modeling and biology, my final contribution to the project has been something between these two groups, curating models of disease risk directly from available literature. I did this, and wrote python programs to tabulate risk from individual trait-o-matic genomes for Prostate Cancer, Type-2 Diabetes, and Ischemic Stroke.
- These models are in a Standard Format, which Fil, Alex and I came up with together. Fil and Alex have since implemented a web interface to upload these models to the site (allowing modularity, so anyone can upload their own model), and print the output as part of their traits-based grouping of trait-o-matic genotypes. Therefore, in addition to seeing all SNPs associated with diabetes in your genome, you would receive a diabetes risk assessment based on those SNPs, based on a published model.
- Finally, I've written another python program to assemble standard-format python programs (above) for simple logistic models: the Logistic Model Generator. It's quite intuitive to use: just run it from the command line, and it will ask you for the rsid's of all relevant SNPs, disease, author name, author name, etc. It will then ask for the odds ratios and confidence intervals for each SNP, and whether the SNP should be modeled as dominant, recessive or multiplicative. From this information it will generate a python program that can be uploaded directly to Fil and Alex's web interface. I like this program since it allows those without a modeling background to transform simple logistic models into python programs readable by the web interface!
- Fil and Alex have now put all of the Models and Model-Generating Program in one place. Thanks guys!
Benefits and Caveats to the Manual Curation Approach:
- The major benefit of this approach is that it enables us to draw, immediately, on all the GWAS studies already performed to predict an individual's disease risk for a number of complex diseases
- Another major benefit is that these studies are peer-reviewed, carefully controlled, and performed by experts in the field, using rational criteria to choose cohorts for analysis; it also includes regular tests of various vital parameters, which could not be the case for a general, population-wide studies. Large repositories of individual genomes cannot, for the foreseeable future, attain this level of manual attention to detail.
- A major caveat is the poor reproducibility of many GWAS studies, which has hurt their reputation, and undermines much of their predictive power
- Another caveat is the fact that, because such studies are done on simple, local cohorts, they may apply effectively to only one population (i.e. white males in finland), and offer poor predictive power for other races, nationalities, etc.
- Due to the above, in cases where diseases have a strong environmental aspect, prediction of absolute risk will be difficult, if not impossible. This is because rates of things like lifetime risk of diabetes can vary many-fold among age- and sex-matched controls for different nations or locales, and it is unclear whether the same SNPs important for diabetes in underweight populations in Finland, would be important for overweight individuals in the US, for instance.
Feeding Manually Curated Disease Risk Models into Trait-O-Matic // 12-06-09
After meeting with Fil, Alex, Anna and Zach last night, we've come up with a general plan for submitting manually curated models to Trait-O-Matic so that, in addition to trait-based SNP reports for individual genomes, you would also be able to see your predicted phenotype (disease risk, height, etc.) for that trait, based on a vetted model from the literature. I think this has some advantages over the extensible methods planned by the modeling and biology groups since, although such a method would be powerful, it would lack the careful oversight provided by peer-reviewed publications in reputable journals.
To make this possible, Alex and Fil are implementing a modification to trait-o-matic that will accept python scripts predicting the disease risk associated with the various SNPs in a genome. I am writing some example scripts for common diseases, to be used by their interface. An example of this, for predicting the risk of Type-2 Diabetes, is shown below; it accepts the two nucleotides given for each SNP in the individual's genome and gives a prediction of overall risk, along with the publication and study that prediction is based on, the PubMed ID of the relevant paper, the method used to calculate the risk, and caveats for interpreting it:
<syntax type="python">
- rsid:1799884
- rsid:1260326
- rsid:560887
- rsid:10830963
- Disease: Diabetes Mellitus, Non-Insulin-Dependent
- Source: Reiling et al. 2009, Dutch New Hoorn Study
- PMID: 19533084
- Model: Genetic Score
- Output: Relative risk of disease by age 53, compared to the general population
- Caveats: Genotype cannot on its own predict diabetes risk, which varies substantially with age, race, BMI and family history.
- Import libraries
import sys
- Check for all inputs
if sys.argv[1].count('x') + sys.argv[2].count('x') + sys.argv[3].count('x') + sys.argv[4].count('x') > 0:
print 'ERROR'
- Calculate overall risk
risk_alleles = sys.argv[1].count('A') + sys.argv[2].count('C') + sys.argv[3].count('G') + sys.argv[4].count('G') if risk_alleles == 0 or risk_alleles == 1:
Risk = .75 Lower = .55 Upper = 1.02
if risk_alleles == 2:
Risk = .78 Lower = .64 Upper = .95
if risk_alleles == 3:
Risk = .89 Lower = .76 Upper = 1.04
if risk_alleles == 4:
Risk = 1.00 Lower = .81 Upper = 1.18
if risk_alleles == 5:
Risk = 1.17 Lower = .97 Upper = 1.41
if risk_alleles == 6 or risk_alleles == 7 or risk_alleles == 8:
Risk = 2.05 Lower = 1.50 Upper = 2.80
print 'RR',Risk,Lower,Upper
</syntax>
If anyone feels left out or wants to contribute something in these final days, just email me; it should be very easy to add additional models to the database now that this framework is in place!
Calculating Absolute Risk of Prostate Cancer // JT 11-22-09
Here are the articles I mentioned by email regarding absolute prostate cancer risk prediction, using both SNPs and family history. The first article is the most useful as a test case (since we can apply it NOW, directly, to the 25 genomes in trait-o-matic, and predict their risk of getting prostate cancer), while the other two are just other studies of multiple SNPs and their combinatorial effects on cancer risk. Have fun at the meeting tonight guys, and on thanksgiving vacation-
- Estimation of Absolute Risk of Prostate Cancer Using Genetic Markers and Family History
- Cumulative Association of Five Genetic Variants with Prostate Cancer
- Utility of Incorporating Genetic Variants for the Early Detection of Prostate Cancer
Other disease-related multi-SNP cases are reported in my 11-01-09 entry.
A Neat Little Toy // JT 11-12-09
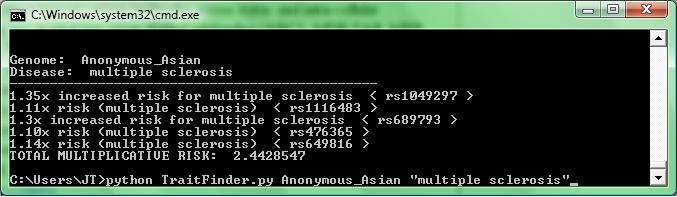
In class I demonstrated my little toy python program for finding all the SNPs containing a given phrase in a given Trait-O-Matic genome, collecting them in one place, and calculating their combined risk (assuming a multiplicative model). The code is Here. Apologies for zipping it; for some reason, OWW gives me an error message when I try to upload .py files. o_O
To find all the SNPs corresponding to a given trait, you just run this program from the command line, and supply both the phrase to search for, and Trait-O-Matic genotype. For example, running:
>>python TraitFinder.py Anonymous_Asian "multiple sclerosis"
Would yield the following output:
I should note that the list of genotypes is on the Trait-O-Matic Website. You can play around with the code to output collections of data, output to text files, etc., but this is just a toy. Fil and Brett are working on properly accessing the Trait-O-Matic datbase, rather than re-parsing it from its output.
I think this can make a good starting-point for building in additional functionality; for instance, it would be relatively straightforward to write a small database of actual logistic regression model parameters (from the literature), then incorporate them into this python script such that once the SNPs are found, they are matched to the database and plugged into real literature models of disease risk.
I've also mentioned logistic regression a bunch of times; probably the best sources are (unsurprisingly) The Wikipedia Page and the old paper I posted on Estimating Prostate Cancer Risk.
A Practical Project Outline // JT 11-10-09
There seems to be a lot of wavering with respect to the project. I've identified what I want my contribution to be, however, so I'm moving right ahead. Here's the overall class project I think makes sense:
- Group the SNPs/traits identified by trait-O-matic and organize into groups. It sounds like Fil and Brett are already on this, fantastic. On its own, this is a fantastic addition to trait-O-matic, and I'm sure not trivial to accomplish.
- Using a handful of grouped SNPs for a single trait (e.g. colon cancer risk), use the simple LD-minimization/multiplicative risk model to predict overall disease risk for each genome on trait-O-matic. Remember that the same model is useful for predicting colon cancer risk (see below) and is used commercially (Navigenics) to make disease risk predictions. We would posit this only as a maximum-ignorance model of interactions, which could be updated as new epigenetic interactions are empirically discovered.
- From the literature, identify a number of "combined SNP/lifestyle/family history" studies for each of a few complex diseases (diabetes, colon cancer, etc.) and extract the relevant parameters needed to quantify risk. Then actually apply this model to each of the genomes available through trait-o-matic. This would determine the amount of work needed to manually curate a number of predictive disease models, and actually apply them within trait-O-matic. As an example, if we limited ourselves to popular logistic regression models, it would take only a few minutes to extract the relevant parameters per paper, and put them in a database.
I'm going to get started on (2) and hopefully demonstrate some very simple results on Thursday.
Modeling SNP Interactions // JT 11-01-09
- Cumulative Association of Five Genetic Variants with Prostate Cancer
- Combination of Multiple Genetic Risk Factors Is Synergistically Associated With Carotid Atherosclerosis in Japanese Subjects With Type 2 Diabetes
- Proof of principle of potential clinical utility of multiple SNP analysis for prediction of recurrent venous thrombosis
- Combining Information from Common Type 2 Diabetes Risk Polymorphisms Improves Disease Prediction
It's clear from these reports that predicting the combined effect of several SNPs will change on a case-by-case basis, and that simple epistasis models (Bliss independence, Loewe additivity) combined with reasonable assumptions about typical epistatic interactions, may not be a practical way to approach gene interactions.
Thoughts on Project Organization // JT&AT 10-30-09
It's good to see the project coming together. In particular, the suggestion to focus on gene interactions, and make the Cupid/heritability a subset of that, makes much more sense than the other way around (i.e. focusing on heritability and making gene interactions a subset). We think there are a few logical sub-projects that clearly stand out here:
- Finding or Constructing a Maximum-Ignorance Model of Genetic Interactions. For example, given 5 SNPs that affect the risk of myocardial infarction in different ways, and NO further information, what upper and lower limits can be placed on the combined risk they imply? Obtaining data to incorporate into this model should initially be easy, since trait-o-matic already pulls risk data from SNPedia (i.e. this database exists already).
- Finding or Constructing a Partial-Ingorance Model of Genetic Interactions. One group could focus on identifying or constructing a model that incorporates familial information and lifestyle into the prediction of disease risk.
- Small-Scale Manual Curation of Genetic Interaction Data for One Disease. One group could perform manual curation of case studies in the literature, like George's combined SNPs for predicting MI risk, both to test the validity of the models above, and to identify the opportunities and pitfalls of incorporating such data into a genetic interaction database, if done on a larger scale.
- Parsing OMIM for Heritability Information. This is necessary for Cupid-type ideas, and would make a nice addition to trait-o-matic in its own right!
HGMD and GeneCards Update // 10-20-09
Anna's page has the writeup for this week, including a nice example of how to cross-reference the information in these databases with OMIM and SNPedia, for instance to predict the possibility of a child inheriting polycystic kidney disease.
SNP-Cupid Project Details // 10-07-09
So, a slightly more substantive look at this "SNPCupid" idea. How would it work in practice? What might it be able to tell us? How could we implement it, as a class, to produce an interesting (and potentially useful?) final project?
I think the best way to do this is to work through one example: male pattern baldness. We chose this as something that might be considered either medical or cosmetic (albeit more the latter than the former, to be sure). We also chose it because it does not fall into one of the more commonly known patterns of inheritance (autosomal dominant, autosomal recessive, sex-linked), but is likely polygenic. This presents a special challenge for attempting to predict phenotype.
We performed a search for "male pattern baldness," (MPB) as well as for its medical synonym - "androgenetic allopecia" - in GeneTests, OMIM and SNPedia. GeneTests unfortunately turned out not to have any information on male patterned baldness, but OMIM and SNPedia had plenty.
SNPedia turned up five relevant SNPs:
- Rs6152 (G) is present in 97% of men with baldness, but only 70% without.
- Rs2223841 (A) increases risk of going bald before age 40.
- Rs1160312 (A), along with mutation in Androgen Receptor (AR), increases risk of baldness 7-fold.
- Rs2180439 (T) increases the risk of MPB 2-fold.
- Rs1385699 (C) is associated with baldness in men in a recent study.
Among these, three were linked directly to loci also catalogued in OMIM, and containing information about heritability, as well as linkage with other genes (which is interesting, but I won't get into it here). Information from NCBI's "genome" section also turned out to be useful. Rs6152 is a SNP in AGA2, while Rs1160312 and Rs2180439 are SNPs in AGA3 (loci catalogued in OMIM).
AGA2 is on the X-chromosome. Its X-linked inheritance pattern, coupled with poor penetrance of MPB in women, means that women may be able to act as carriers for the relevant MPB gene. We also know that the Rs6152(G) SNP appears in 97% of bald men, but only 70% of non-bald men; males not carrying this SNP may
therefore be significantly less likely to go bald. Because AGA2 is X-linked, these men would necessarily pass this gene to their daughters, who could potentially act as carriers.
AGA3 is on (autosomal) chromosome 20. Together with an AR mutation, the two SNPs at this locus have up to a 14-fold impact on the risk of going bald.
How might this kind of information be useful? Since SNPs are relatively straightforward to test for, they are a reasonable starting point for any method that hopes to provide personal genetic information. Coupled with the OMIM data, we can also learn something about their heritability. A mother carrying the Rs6152(G) SNP, would have a 50% chance of passing this on to her male child, which would then impact his risk of going bald. Likewise, existence of the Rs1160312(A) SNP in both parents would suggest a significant probability of the child inheriting at least one of the associated alleles (and therefore of having a strongly increased risk for MPB).
If we wanted to systematically extract this sort of information for hundreds of traits/diseases, could we? My intuition is that this would plausible for traits with simple inheritance patterns (i.e. autosomal recessive), while descriptions may be slightly too ramshackle both in OMIM and SNPedia to expect a computer to consistently extract meaningful information on polygenic traits (though this would make for an interesting challenge). That said, the above example illustrates the kinds of information available from these resources, and how they can be used together to make predictions about both parental and F1 genotypes and phenotypes.
How to move forward: Focusing on a set of genes with well-know inheritance patterns (i.e. autosomal-recessive), a reasonably simple program could be written in python to identify SNPs for a trait/disease, cross-reference them to OMIM, and pull down inheritance information. It would need to parse the OMIM data and look for high-quality indications that the mode of inheritance is either autosomal recessive, dominant, etc. (by "high-quality," I mean there would have to be some way to score whether what OMIM says about a certain gene's inheritance pattern is reliable or not). Once this is done, a set of dummy genomes could be constructed to test the ability of the program to predict their offspring's genotypes and phenotypes. In fact, real data can be used to construct these dummy genomes by referencing the studies in which the SNPs were first identified (I won't get into details here, but I think this would make for a much-needed sanity check). This sounds like a fun challenge to me, so I'm up for it.
Project Proposals // 09-29-09
In the course of the last few lectures, two concepts have stuck out for me:
- While a long way off, direct genomic manipulation of humans, whether for medical or cosmetic purposes, is likely inevitable (and indeed, primitive genetic therapies do exist)
- The continued survival/expansion of the human race will require intelligent human planning and intervention
First Idea: while human genomic modification is difficult, we have had very good success engineering microbial genomes (and are probably on the cusp of assembling them from scratch). Since the human organism is really an amalgamation of both homo sapiens and the millions of commensal skin, mouth and gut flora that live on or in us (and are responsible for various vital functions, such as vitamin biosynthesis), a good way of making a Human 2.0 is not by modifying our own genomes, but by those of our indigenous microbes! An idea for a Human 2.0 project could therefore be along the lines of, "How can we reengineer our endogenous microbiota to improve human health?"
Second Idea: This ties in with our logistic equation plotting assignment two weeks ago. The logistic equation is often used in population/evolutionary biology to describe the growth of organisms on some fixed set of resources, which determines the "carring capacity," or maximal number of organisms that can exist on this fixed set of resources. If the resources increase, the population can do the same; if it goes down, the population must follow. A (tragic) human example of this behavior occurred in 1845 with the onset of the Irish Potato Famine, where the sudden, dramatic loss of potatoes (the major Irish source of carbon, among other nutrients) caused a sudden, dramatic drop in population size (incidentally, it dropped to around the size of the population prior to the introduction of potatoes! Refs to be added.).
Right now, humans rely on a number of resources which appear abundant (good soil, good air, potable water, oil and natual gas reserves), but are actually being consumed faster than they are replenished. Assuming we were to switch only to renewable energy/food/water/soil sources, however, by what % would the human population have to drop? What new technologies would be necessary to maintain our current 6 billion people? Are there fundamental limits on how big this carrying capacity can become, and are we already above them? Essentially I'm asking: Is it possible to calculate the human carrying capacity on earth? How large can such a carrying capacity be on earth? And are there fundamental limits to it? (incident sunlight for energy, physical space on earth, timescales for waste remediation, etc.)
p53 Sequence Analysis Assignment // 09-24-09
Here's my code from assignment 3. As it was essentially my first time with Python, I didn't bother to learn functions and just sort of copy/pasted the reused code; additionally, I've never really done any string manipulation before, so the functions I used were those google recommended to me; with these two factors combined, the code is very long and unwieldly. Heartfelt apologies :P
<syntax type="python">
- -- Biophysics 101 Assignment 3, Due 09/24/09, J.P. Torella --
- -- Import needed modules --
from string import * from re import * from random import * from numpy import * import matplotlib.pyplot as plt
- -- I've pasted the p53 sequence into a file called p53.seq. I'll have the program read it into a string --
filename = "p53.seq" infile = open(filename,"r") data = "default" seq = "" while (data != ""):
data = infile.readline() seq += rstrip(data) # necessary to use rstrip to remove return char. from end of ea. line
print "\nThe Sequence Is:" print seq infile.close()
- -- Part 1: Determine GC content --
Gs = seq.count("g") Cs = seq.count("c") GC_Content = 100*float(Gs + Cs)/float(len(seq)); print "\nThe GC Content Is (%):" print GC_Content
- -- Part 2: Determine the reverse complement --
Reversed_Sequence = seq[::-1] in_table = "atgc" out_table = "tacg" translation_table = maketrans(in_table, out_table) Reverse_Complement = Reversed_Sequence.translate(translation_table) print "\nThe Reverse Complement Is:" print Reverse_Complement
- -- Part 3: Protein sequences from all reading frames --
total = 1020 position = 1 remainder = total - (position-1) AA_Sequence = "" while (remainder >= 3):
AA = "Z" temp_seq = seq[(position-1):position+2] if (temp_seq == "ttt") or (temp_seq == "ttc"): AA = "F" if (temp_seq == "tta") or (temp_seq == "ttg") or (temp_seq == "ctt") or (temp_seq == "ctc") or (temp_seq == "cta") or (temp_seq == "ctg"): AA = "L" if (temp_seq == "tct") or (temp_seq == "tcc") or (temp_seq == "tca") or (temp_seq == "tcg") or (temp_seq == "agt") or (temp_seq == "agc"): AA = "S" if (temp_seq == "cgt") or (temp_seq == "cgc") or (temp_seq == "cga") or (temp_seq == "cgg") or (temp_seq == "aga") or (temp_seq == "agg"): AA = "R" if (temp_seq == "cct") or (temp_seq == "ccc") or (temp_seq == "cca") or (temp_seq == "ccg"): AA = "P" if (temp_seq == "ggt") or (temp_seq == "ggc") or (temp_seq == "gga") or (temp_seq == "ggg"): AA = "G" if (temp_seq == "gct") or (temp_seq == "gcc") or (temp_seq == "gca") or (temp_seq == "gcg"): AA = "A" if (temp_seq == "gtt") or (temp_seq == "gtc") or (temp_seq == "gta") or (temp_seq == "gtg"): AA = "V" if (temp_seq == "act") or (temp_seq == "acc") or (temp_seq == "aca") or (temp_seq == "acg"): AA = "T" if (temp_seq == "att") or (temp_seq == "atc") or (temp_seq == "ata"): AA = "I" if (temp_seq == "taa") or (temp_seq == "tga") or (temp_seq == "tag"): AA = "*" if (temp_seq == "tat") or (temp_seq == "tac"): AA = "Y" if (temp_seq == "tgt") or (temp_seq == "tgc"): AA = "C" if (temp_seq == "cat") or (temp_seq == "cac"): AA = "H" if (temp_seq == "caa") or (temp_seq == "cag"): AA = "Q" if (temp_seq == "aat") or (temp_seq == "aac"): AA = "N" if (temp_seq == "aaa") or (temp_seq == "aag"): AA = "K" if (temp_seq == "gat") or (temp_seq == "gac"): AA = "D" if (temp_seq == "gag") or (temp_seq == "gaa"): AA = "E" if (temp_seq == "tgg"): AA = "W" if (temp_seq == "atg"): AA = "M" remainder = remainder - 3 position = position + 3 AA_Sequence += AA
print "\nThe +1 Amino Acid Sequence Is:" print AA_Sequence print AA_Sequence.count("*")
- -- Part 4a: Introduce random mutations into the +1 frame at a rate of 1%, and document AA changes --
Amut = "tgc" Tmut = "agc" Gmut = "atc" Cmut = "atg" mut_seq = ""
for i in range(len(seq)):
rand_num = random.random() if (rand_num <= .01): #-- 1% chance of undergoing a mutation
rand_2 = random.random() #-- This will tell us which base the mutation will produce
if (seq[i] == "a"):
if (rand_2 <= .333): mut_seq += Amut[0]
if (rand_2 > .333) and (rand_2 <= .667): mut_seq += Amut[1]
if (rand_2 > .667) and (rand_2 <= 1): mut_seq += Amut[2]
if (seq[i] == "t"):
if (rand_2 <= .333): mut_seq += Tmut[0]
if (rand_2 > .333) and (rand_2 <= .667): mut_seq += Tmut[1]
if (rand_2 > .667) and (rand_2 <= 1): mut_seq += Tmut[2]
if (seq[i] == "g"):
if (rand_2 <= .333): mut_seq += Gmut[0]
if (rand_2 > .333) and (rand_2 <= .667): mut_seq += Gmut[1]
if (rand_2 > .667) and (rand_2 <= 1): mut_seq += Gmut[2]
if (seq[i] == "c"):
if (rand_2 <= .333): mut_seq += Cmut[0]
if (rand_2 > .333) and (rand_2 <= .667): mut_seq += Cmut[1]
if (rand_2 > .667) and (rand_2 <= 1): mut_seq += Cmut[2]
if (rand_num > .01):
mut_seq += seq[i]
total = 1020 position = 1 remainder = total - (position-1) AA_Sequence = "" while (remainder >= 3):
AA = "Z" temp_seq = mut_seq[(position-1):position+2] if (temp_seq == "ttt") or (temp_seq == "ttc"): AA = "F" if (temp_seq == "tta") or (temp_seq == "ttg") or (temp_seq == "ctt") or (temp_seq == "ctc") or (temp_seq == "cta") or (temp_seq == "ctg"): AA = "L" if (temp_seq == "tct") or (temp_seq == "tcc") or (temp_seq == "tca") or (temp_seq == "tcg") or (temp_seq == "agt") or (temp_seq == "agc"): AA = "S" if (temp_seq == "cgt") or (temp_seq == "cgc") or (temp_seq == "cga") or (temp_seq == "cgg") or (temp_seq == "aga") or (temp_seq == "agg"): AA = "R" if (temp_seq == "cct") or (temp_seq == "ccc") or (temp_seq == "cca") or (temp_seq == "ccg"): AA = "P" if (temp_seq == "ggt") or (temp_seq == "ggc") or (temp_seq == "gga") or (temp_seq == "ggg"): AA = "G" if (temp_seq == "gct") or (temp_seq == "gcc") or (temp_seq == "gca") or (temp_seq == "gcg"): AA = "A" if (temp_seq == "gtt") or (temp_seq == "gtc") or (temp_seq == "gta") or (temp_seq == "gtg"): AA = "V" if (temp_seq == "act") or (temp_seq == "acc") or (temp_seq == "aca") or (temp_seq == "acg"): AA = "T" if (temp_seq == "att") or (temp_seq == "atc") or (temp_seq == "ata"): AA = "I" if (temp_seq == "taa") or (temp_seq == "tga") or (temp_seq == "tag"): AA = "*" if (temp_seq == "tat") or (temp_seq == "tac"): AA = "Y" if (temp_seq == "tgt") or (temp_seq == "tgc"): AA = "C" if (temp_seq == "cat") or (temp_seq == "cac"): AA = "H" if (temp_seq == "caa") or (temp_seq == "cag"): AA = "Q" if (temp_seq == "aat") or (temp_seq == "aac"): AA = "N" if (temp_seq == "aaa") or (temp_seq == "aag"): AA = "K" if (temp_seq == "gat") or (temp_seq == "gac"): AA = "D" if (temp_seq == "gag") or (temp_seq == "gaa"): AA = "E" if (temp_seq == "tgg"): AA = "W" if (temp_seq == "atg"): AA = "M" remainder = remainder - 3 position = position + 3 AA_Sequence += AA
print "\nThe Mutated Amino Acid Sequence Is:" print AA_Sequence
- -- Part IV B: Number of stop codons in mutated sequences?
stop_codons = zeros((1000,1))
for j in range(1000):
Amut = "tgc" Tmut = "agc" Gmut = "atc" Cmut = "atg" mut_seq = ""
for i in range(len(seq)):
rand_num = random.random()
if (rand_num <= .01): #-- 1% chance of undergoing a mutation
rand_2 = random.random() #-- This will tell us which base the mutation will produce
if (seq[i] == "a"):
if (rand_2 <= .333): mut_seq += Amut[0]
if (rand_2 > .333) and (rand_2 <= .667): mut_seq += Amut[1]
if (rand_2 > .667) and (rand_2 <= 1): mut_seq += Amut[2]
if (seq[i] == "t"):
if (rand_2 <= .333): mut_seq += Tmut[0]
if (rand_2 > .333) and (rand_2 <= .667): mut_seq += Tmut[1]
if (rand_2 > .667) and (rand_2 <= 1): mut_seq += Tmut[2]
if (seq[i] == "g"):
if (rand_2 <= .333): mut_seq += Gmut[0]
if (rand_2 > .333) and (rand_2 <= .667): mut_seq += Gmut[1]
if (rand_2 > .667) and (rand_2 <= 1): mut_seq += Gmut[2]
if (seq[i] == "c"):
if (rand_2 <= .333): mut_seq += Cmut[0]
if (rand_2 > .333) and (rand_2 <= .667): mut_seq += Cmut[1]
if (rand_2 > .667) and (rand_2 <= 1): mut_seq += Cmut[2]
if (rand_num > .01):
mut_seq += seq[i]
total = 1020
position = 1
remainder = total - (position-1)
AA_Sequence = ""
while (remainder >= 3):
AA = "Z"
temp_seq = mut_seq[(position-1):position+2]
if (temp_seq == "ttt") or (temp_seq == "ttc"): AA = "F"
if (temp_seq == "tta") or (temp_seq == "ttg") or (temp_seq == "ctt") or (temp_seq == "ctc") or (temp_seq == "cta") or (temp_seq == "ctg"): AA = "L"
if (temp_seq == "tct") or (temp_seq == "tcc") or (temp_seq == "tca") or (temp_seq == "tcg") or (temp_seq == "agt") or (temp_seq == "agc"): AA = "S"
if (temp_seq == "cgt") or (temp_seq == "cgc") or (temp_seq == "cga") or (temp_seq == "cgg") or (temp_seq == "aga") or (temp_seq == "agg"): AA = "R"
if (temp_seq == "cct") or (temp_seq == "ccc") or (temp_seq == "cca") or (temp_seq == "ccg"): AA = "P"
if (temp_seq == "ggt") or (temp_seq == "ggc") or (temp_seq == "gga") or (temp_seq == "ggg"): AA = "G"
if (temp_seq == "gct") or (temp_seq == "gcc") or (temp_seq == "gca") or (temp_seq == "gcg"): AA = "A"
if (temp_seq == "gtt") or (temp_seq == "gtc") or (temp_seq == "gta") or (temp_seq == "gtg"): AA = "V"
if (temp_seq == "act") or (temp_seq == "acc") or (temp_seq == "aca") or (temp_seq == "acg"): AA = "T"
if (temp_seq == "att") or (temp_seq == "atc") or (temp_seq == "ata"): AA = "I"
if (temp_seq == "taa") or (temp_seq == "tga") or (temp_seq == "tag"): AA = "*"
if (temp_seq == "tat") or (temp_seq == "tac"): AA = "Y"
if (temp_seq == "tgt") or (temp_seq == "tgc"): AA = "C"
if (temp_seq == "cat") or (temp_seq == "cac"): AA = "H"
if (temp_seq == "caa") or (temp_seq == "cag"): AA = "Q"
if (temp_seq == "aat") or (temp_seq == "aac"): AA = "N"
if (temp_seq == "aaa") or (temp_seq == "aag"): AA = "K"
if (temp_seq == "gat") or (temp_seq == "gac"): AA = "D"
if (temp_seq == "gag") or (temp_seq == "gaa"): AA = "E"
if (temp_seq == "tgg"): AA = "W"
if (temp_seq == "atg"): AA = "M"
remainder = remainder - 3
position = position + 3
AA_Sequence += AA
stop_codons[j] = AA_Sequence.count("*")
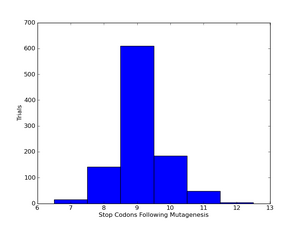
plt.hist(stop_codons,[6.5,7.5,8.5,9.5,10.5,11.5,12.5]) plt.xlabel('Stop Codons Following Mutagenesis') plt.ylabel('Trials') plt.show()
</syntax>
First Assignments // 09-17-09
I just realized that, oddly enough, when I click on "talk" on the main site, versus "talk" on the people page, I end up at different places. So, apparently I haven't actually updated anything yet.
For good measure, here's the second assignment data on the number of stop codons present in the +1 frame, following mutagenesis of the p53 sequence with a frequency of 1%:
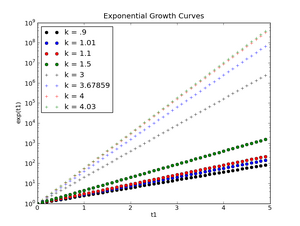
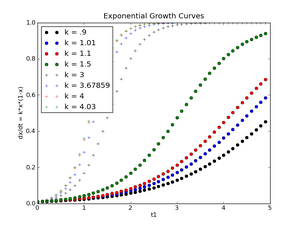
And here's the first assignment data (logarithmic scale for plotting the exponential function, linear scale for plotting the logistic function):