Tianjin protocol
<html> <head> <meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> <title>无标题文档</title> <style type="text/css">
body {
display:block;
background-color: #ffffff;
}
.ys01 {
text-align: left;
}
/* --the header css -- */
- apDiv1 {
position:relative;
float:left;
left:0px; top:0px;
margin-right:0px;
width:1000px; height:350px; z-index:1;
box-shadow: 0px 5px 8px rgba(0,0,0,0.4);
overflow: visible; text-align: left; background-image: url(http://openwetware.org/images/3/37/Header_tianjin.png);
}
/* -- navigator css -- */
- apDiv2 {
position:relative; float:left;
left:0px;
width:1000px; height:40px; z-index:2; font-family: Georgia, "Times New Roman", Times, serif; text-align: left; background-image: url(http://openwetware.org/images/thumb/7/76/Tianjin_index02.jpg/800px-Tianjin_index02.jpg);
}
- apDiv2 table {
font-size: 18px;
text-align: center;
}
.ys01 #apDiv2 table {
text-align: center;
}
.ys01 #apDiv2 table tr td a { color: #FFF;
}
/* -- navigator css -- */
- navDiv li { margin: 0; padding: 0;}
- navDiv ul {margin: 0; padding: 0; list-style: none; z-index:99;}
- navDiv a {text-decoration: none;margin: 0; padding: 0;}
- navDiv
{
/* -- background body of navigator css -- */
position: relative;
float: left;
height: 40px; width:1000px; left: 0px; background-color: #ffffff; background-repeat: repeat-x; box-shadow: 0px 5px 8px rgba(0,0,0,0.4); margin-bottom:40px;
z-index:60;
font-family: Georgia, "Times New Roman", Times, serif;
text-align: left;
}
- navDiv > ul > li {
float: left; margin-left: 30px; position: relative;
}
/* -- when mouse not on the navigator css -- */
- navDiv > ul > li > a {
color: #333333; font-family: Verdana, 'Lucida Grande'; font-size: 15px; line-height: 40px; padding: 11px 20px;
-webkit-transition: color .15s;
-moz-transition: color .15s;
-o-transition: color .15s;
transition: color .15s;
}
/* -- when mouse on the navigator css -- */
- navDiv > ul > li > a:hover {color: #ffffff; background-color: #000000;}
/* -- top and bottom of the sub meun css -- */
- navDiv > ul > li > ul
{
opacity: 0; visibility: hidden; padding: 16px 0 20px 0; background-color: rgb(250,250,250); text-align: left; position: absolute; top: 30px; left: 50%; margin-left: -90px; width: 180px;
-webkit-transition: all 0.3s 0.1s;
-moz-transition: all 0.3s 0.1s;
-o-transition: all 0.3s 0.1s;
transition: all 0.3s 0.1s;
-webkit-border-radius: 5px;
-moz-border-radius: 5px;
border-radius: 5px;
-webkit-box-shadow: 0px 1px 3px rgba(0,0,0,0.4);
-moz-box-shadow: 0px 1px 3px rgba(0,0,0,0.4);
box-shadow: 0px 1px 3px rgba(0,0,0,0.4);
}
- navDiv > ul > li:hover > ul
{
opacity: 1; top: 40px; visibility: visible;
}
- navDiv > ul > li > ul:before
{
content: ; display: block; border-color: transparent transparent rgb(250,250,250) transparent; border-style: solid; border-width: 10px; position: absolute; top: -20px; left: 50%; margin-left: -10px;
}
- navDiv > ul ul > li { position: relative;}
/* -- when mouse not on the submeun css -- */
- navDiv ul ul a
{
color: rgb(50,50,50); font-family: Verdana, 'Lucida Grande'; font-size: 13px; background-color: rgb(250,250,250); padding: 5px 8px 7px 16px; display: block;
-webkit-transition: background-color .1s;
-moz-transition: background-color .1s;
-o-transition: background-color .1s;
transition: background-color .1s;
}
- navDiv ul ul ul
{
visibility: hidden; opacity: 0; position: absolute; top: -16px; left: 206px; padding: 16px 0 20px 0; background-color: rgb(250,250,250); text-align: left; width: 160px;
-webkit-transition: all .3s;
-moz-transition: all .3s;
-o-transition: all .3s;
transition: all .3s;
-webkit-border-radius: 5px;
-moz-border-radius: 5px;
border-radius: 5px;
-webkit-box-shadow: 0px 1px 3px rgba(0,0,0,.4);
-moz-box-shadow: 0px 1px 3px rgba(0,0,0,.4);
box-shadow: 0px 1px 3px rgba(0,0,0,.4);
}
- navDiv ul ul > li:hover > ul { opacity: 1; left: 196px; visibility: visible;}
/* -- when mouse on the navigator css -- */
- navDiv ul ul a:hover
{
background-color: #777777; color: rgb(240,240,240);
}
/* Back to top css */
.back-to {
position: fixed; bottom: 35px; *bottom: 50px; _bottom: 35px; right: 0px; z-index: 999; width: 50px; zoom: 1; }
- html .back-to {
/* 不能用 _position 这种写法,因为它在IE7+也会执行expression。。。 */
position: expression(function(ele){ele.runtimeStyle.position='absolute';Expressions.style.position.fixed(ele);}(this))
}
.back-to {
float: right; display: block; width: 50px; height: 50px; background: url(http://a.xnimg.cn/imgpro/button/back-home.png) no-repeat 0 0; outline: 0 none; text-indent: -9999em;
}
.back-to:hover {
background-position: -50px 0 }
.back-to .back-top {
float: right; display: block; width: 50px; height: 50px; background: url(http://a.xnimg.cn/imgpro/button/back-top.png) no-repeat 0 0; margin-left: 10px; outline: 0 none; text-indent: -9999em; }
.back-to .backtotop {
float: left; display: block; width: 50px; height: 50px; background: #666 url(http://a.xnimg.cn/imgpro/arrow/btt.png) 8px -57px no-repeat; margin-bottom: 15px; outline: 0 none; text-indent: -9999em; -moz-border-radius: 4px; -khtml-border-radius: 4px; -webkit-border-radius: 4px; border-radius: 4px; position: relative; -moz-box-shadow: 0px 0px 15px #ccc; -webkit-box-shadow: 0px 0px 15px #ccc; box-shadow: 0px 0px 15px #ccc; }
.back-to .backtotop:hover {
background-color: #333; background-position: 8px 13px; }
.back-to .backtotop .back-tip {
position: absolute; visibility: hidden; top: -31px; left: -10px; }
.back-to .backtotop:hover .back-tip {
visibility: visible; }
.back-to .back-top:hover {
background-position: -50px 0; }
/* --video css -- */
- apDiv3 table {
text-align: center;
}
.ys01 #apDiv6 table { text-align: left;
}
.ys02 {
font-size: 36px;
color: #F00;
}
- apDiv10 {
position:relative;
float:left;
display:block;
left:0px;
width:961px; z-index:3;
box-shadow: 0px 5px 8px rgba(0,0,0,0.4);
text-align: left; font-family: Georgia, "Times New Roman", Times, serif; font-size: 18px; color: #000; background-color: #ffffff;
padding: 20px;
}
/*hidden section*/
.firstHeading{display:none;}
- sidebar-main{display:none;}
- p-cactions{display:none;}
- p-personal{display:none;}
</style> </head>
<body class="ys01";>
<a stats="site_footer_back_to_top" class="backtotop" href="#top" onclick="if(Sizzle('#sidebar2 .ready-to-fix')[0]) Sizzle('#sidebar2 .ready-to-fix')[0].style.position = 'static';window.scrollTo(0,0);if(Sizzle('#sidebar2 .ready-to-fix')[0]) Sizzle('#sidebar2 .ready-to-fix')[0].style.position = ;return false;">Back to top <img stats="site_footer_back_to_top" src="http://openwetware.org/images/a/a7/TJU2012-Back-tip.png" class="back-tip"> </a>
</html>
Buffer Preparation
(1)5×TBE: Add 54g Tris Base, 3.7224g EDTA Na2﹒2H2O and 27.5g borieacid in a 1000mL volumetric flask. Add D.D. water until the final volume is 1000 mL. Make the pH into 8.25~8.30.
(2)1×TBE: Add 200mL of 5×TBE in a 1000mL volumetric flask. Add D.D. water until the final volume is 1000 mL.
(3)50×TAE: Add 242g Tris Base, 23g EDTA and 57.1mL A cetic acid glacial in a 1000mL volumetric flask. Add D.D. water until the final volume is 1000 mL.
(4)1×TAE: Add 20mL of 50×TAE in a 1000mL volumetric flask. Add D.D. water until the final volume is 1000 mL.
Gel Pouring
PAGE Gel(10%)
| Content | Volume |
|---|---|
| Acrylamide Gel 30% | 15ml |
| 5×TBE | 6ml |
| ddH2O | 9ml |
| TEMED | 20µl |
| ASP | 150µl |
| Total | 15ml |
Prepare gel according the volumes in the table above
Pour gel into casting tray, and insert comb (make sure there are no bubbles)
Put the whole setup in 37℃ incubator for 30 min
Fill in TBE-Buffer
Prepare the sample by adding 6×loading buffer containing Golden View
Agarose Gel
1.Place 1.0(or 2.0) g of agarose in a clean 600 mL beaker on a scale.
2.Prepare the 1×TAE buffer as described.
3.Add 100 mL of 1×TAE buffer.
4.Microwave for 2 minutes.
5.If the solution is not clear, swirl and microwave for another 15 seconds.
6.Cool the beaker in an ice water bath. Keep swirling the beaker to prevent polymerization. Add the thermometer and keep checking the temperature.
7.When the solution reaches 60°C, remove it from the ice water bath.
8.Add 5µL of EB.
9.Pour the contents of the beaker into the gel cassette and insert the comb.
10.Use a glass Pasteur pipet to push any bubbles to the corners of the gel.
11.Leave the cassette covered and let the assembly sit for thirty minutes to complete ensure complete polymerization.
Origimi
Design
We want to design a two-dimensional DNA origami plate which can be folded by our locker into a roll. The dimensions of the origami plate are properly selected so the drug stays encapsulated inside it. The final dimensions are the following:
Length: 50 nm
Width: 35 nm
Thickness: 2 nm
To enable the origami to fold into a roll, the three arm locker should be combined with it. The staple strands in two corners of the origami plate are extended with the sequences of strand A, and the staple strands in the other two corners are extended with the sequences of strand C.
We used the cadnano 2 program for our design.
The scaffold used in this design is M13mp18 single strand DNA with 7249 nt, from New England Biolabs LTD. The desired structure and dimensions are achieved with the help of two types of staples:
core: 58 staples give stability to the whole structure.
edge: 4 staples give stability to the edges and with extending sequences of A or C.
Folding and Characterization
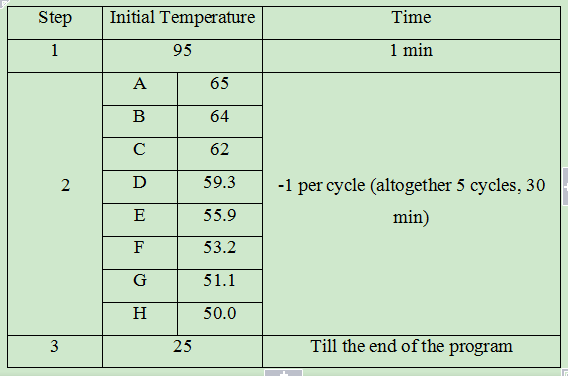
The mixture of scaffold and different staples were subjected to a thermal annealing process composed of 3 steps that allowed the folding. Different initial temperatures varying from 50.0 to 65.0 in step 2 were tested. Electrophoresis showed that low initial temperatures in step 2 have relatively high origami yield.
Different Thermal Annealing Processes of DNA Origami
Formation of the Hairpins
A, B and C did a snap-cooling procedure to form hairpin structure: heating at 90 ◦C for 5 minutes and cooling on ice for 1 minute. The hairpins were allowed to equilibrate at room temperature for 30 minutes before use. System including: 60µL ddH2O, 30µL 10µM DNA(A,B,C),10µL 10×Reaction Buffer(RB) total volume is 100µL/tube. (3µM reaction) For 6µM I: 30µL ddH2O, 60µL 10µM DNA,10µL 10×Reaction buffer
Assembly of the Three-arm Locker
Dilution oligonucleotides
Add water to 100µM. Then 10µL 100µM DNA+90µL water are mixed to get 10µM DNA.
Make A, B and C form hairpins as described.
Make A, B and C form hairpins as described.
Assembly(conc:750nM)
(1)single:7.5μL hairpin A+22.5μL 1xRB, total vol 30μL
(2)assemble: 7.5μL hairpin A+7.5μL hairpin B+7.5μL hairpin C+ 6μM I 7.5μL, total vol 30μL
(3)leakage: 7.5μL hairpin A+7.5μL hairpin B+7.5μL hairpin C+ 7.5μL 1xRB, total vol 30μL
(4)annealed: 7.5μL hairpin A+7.5μL hairpin B+7.5μL hairpin C+ 7.5μL 1xRB (95◦C for 5 minutes first) , total vol 30μL
(5)(ABCI2×):7.5μL hairpin A+7.5μL hairpin B+7.5μL hairpin C+ 6μM I 7.5μL(95◦C for 5 minutes first), total vol 30μL
Preparation of gold-DNA nanoconjugates
1、Prepare 10 mM TCEP.
2、 Pipette 8μL of 500 mM DNA into a microcentrifuge tube
3、Add 1μL of 500 mM acetate buffer (pH 5.2) and 1μL of 10 mM TCEP to the tube to activate the thiol-modified DNA. Incubate the sample at room temperature for 1 h.
4、Transfer 56μL of the gold nanoparticles and 1334ul H20 to the tube, and then add the TCEP-treated thiol DNA with gentle shaking by hand.
5、Cap the tube and store it in a drawer at room temperature for at least 16 h. Magnetic stirring may also be applied to facilitate the reaction.
6、 Add 14μL of 500 mM Tris acetate (pH 8.2) buffer dropwise to the tube with gentle hand shaking.
7、Add 14μL of 1 M NaCl dropwise to the tube with gentle hand shaking. Store the tube in a drawer for at least another day before use.
8、Centrifuge the two tubes at 1500g at room temperature (23–25 °C) on a centrifuge for 60 min.
9、Remove the tube from the centrifuge. The supernatant should be clear, and the nanoparticles should be at the bottom of the tubes. If a red color can still be observed in the supernatant, centrifuge for another 5 min.
10、Gently pipette off as much supernatant as possible to remove free DNA. Again, disperse the nanoparticles in 30μL H2O.
11、 The resulting Au-DNA conjugates were loaded on 2% agarose gels and run for one hour at 100 V.
<html>
</body> <html>