Shivum Desai Journal Week 11
Scientific Journal Week 11 (Continuation of Week 10 Assigment)
Purpose
Purpose of this weeks joural is to continue the steps required to finish the week 10 assignment. This inclucded further research if necessary into our projects and then the formation of a powerpoint for presentation.
Hypothesis
It is hypothesized that the rapid progressor subjects will have a greater number of amino acid mutations when comparing their first visit to their last. It is also hypothesized that the there will be a correlation, negative or positive, between amino acid mutations and CD4 cell count.
Methods
1. The first step was to reselect the subjects for out experiment.
2. Next I performed a multiple sequences alignment with all subjects clones from first and last visits. This data was then analyzed to find a pattern in the amino acid changes.
3. After this the sequences from the Huang paper was used to compare to the Markham sequences using a protein rendering and analysis program. The purpose behind this was to find the differences between the Huang paper and the Markham sequences.
4. We then reanalyzed a box shade graph to count the number of amino acid mutations that were present in all these subject. The results showed several more mutations than previously because the previously counted mutations were off of virus sequences that were not complete.
Week 11:
5. The next step was to perform a cluster alignment of all the sequences we are using to show the difference between each subject's first and last visit.
- The distances were recorded for each subject.
6. Then a region of mutational clustering was found and connections and ideas as to why this region contains so many mutations was formulated.
7. Cytokines were researched and a conclusion was drawn.
8. Presentation was designed
Data/Files
Experimental Subjects
Nonprogressor
- Subject 13-visits 1 and 5
- Subject 12-visits 1 and 8
Rapid Progressors
- Subject 10-vists 1 and 6
- Subject 4-visits 1 and 4
Protein Structure Analysis

- This structure was used to generate the Markham sequences to be compared to experimental subject sequences for comparison.
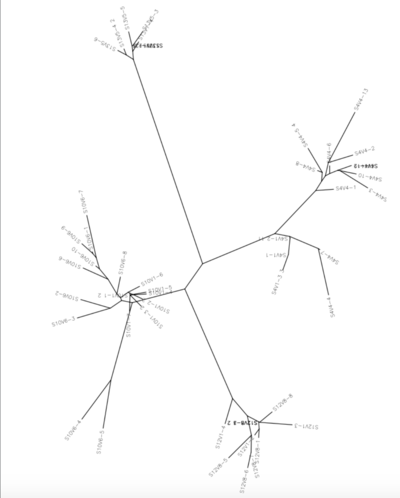
ClustalDist Results
- Following data is the distance between each subject's first and last visit
- Subject 10: 0.116
- Subject 4: 0.117
- Subject 12: 0.053
- Subject 13: 0.021
(10&4 are rapid progressors/12&13 are non progressors)
Analysis of Amino Acid Mutations
- After analyzing the amino acid changes for each subject's first and last visits, there was no observable pattern in the changes of amino acids. In other words, there was no common observable pattern of change of an amino acid in a subject's first visit sample compared to his last visit. There were observable mutations that occurs, but it did not appear that there was any connection (i.e unpolar--> polar or charged-->unpolar) between them.
- Not all subject had amino acid changes comparing from first visit sample to last.
- 30% of all aa mutations were found to be polar to hydrophobic or hydrophobic to hydrophobic
Amino Acid Mutations Between Subject's First and Last Visits
- Subject 4: 4 mutation
- Subject 10: 2 mutations
- Subject 12: 2 mutations
- Subject 13: 01mutations
Greater than 50% of mutations were found in region from amino acid 38 to 42.
Presentation Link
Scientific Conclusion
After discussing this project with Dr. Dahlquist, a new set of subjects was selected in order to have an even number of rapid progresses and non progressors. This way, when we proved or disproved our hypothesis, our data would be sustainable. We then performed a multiple sequence alignment with all subjects and their respective visits and clones. We used clones from each subjects first and last visits. The multiple sequences alignment showed no pattern of amino acid changes when comparing each subject's first visit amino acid sequences to their last visit amino acid sequences. The next step was to generate a protein structure using a amino acid sequences form the Huang article. This was then compared to the subjects that we are using to find several amino acid differences amongst the sequences, but nothing extremely different. The unrooted tree present also shows the genetic difference between each of the four subjects. The results of the ClustalDist also showed that the rapid progressor subjects had a larger difference between their first and last visits than the non progressor subjects did. This is in support of our hypothesis, that rapid progressor subjects will have larger amount of mutations between their first and last visits. However, when looking at the number of amino acid mutations that occurred across all the subject, not all the subjects had large amounts of amino acid changes. Subjects 4 and 13 showed almost no amino acid mutations when comparing the clones of their first and last visits, while subjects 10 and 12 did show several mutations. This is interesting, because it disproves our hypothesis, that the rapid progressor subject would have a great number of amino acid mutations, which is contrary to what the ClustalDist information supports.
Week 11 Cont. The amino acid region from 38-42 was found to have the highest number of mutations. This region is also present in the V3 loop area which is were polar cytokine receptor molecules attach. This led us to believe that this region is meant to be the variable region for cytokine attachment. Which is why there are so many mutations in this area, because the virus would need to mutate most frequently here in order to avoid detection by the body's autoimmune response. Additionally, the rapid progressor viruses would have the most amount of mutations because they would need to adapt and mutate the most often to avoid the autoimmune response. Thus, these viruses are going to have the greatest amount of mutations.
Useful Links
- Shivum Desai
- Courtney Merriam
- Week 11 Assignment Page
- Week 11 Shared Journal Page
- Template:Shivum A Desai
References
- Kirchherr, J. L., Hamilton, J., Lu, X., Gnanakaran, S., Muldoon, M., Daniels, M., Kasongo, W., Chalwe, V., Mulenga, C., Mwananyanda, L., Musonda, R.M., Yuan, X., Montefiori, D.C., Korber, M.T., Haynes, B.F., & Musonda, R. M. (2011). Identification of amino acid substitutions associated with neutralization phenotype in the human immunodeficiency virus type-1 subtype C gp120. Virology, 409(2), 163-174. DOI: 10.1016/j.virol.2010.09.031
- Markham, R.B., Wang, W.C., Weinstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. dos: 10.1073/pnas.95.21.12568
- Yonezawa, A., Hori, T., Takaori-Kondo, A., Morita, R., & Uchiyama, T. (2001). Replacement of the V3 Region of gp120 with SDF-1 Preserves the Infectivity of T-Cell Line-Tropic Human Immunodeficiency Virus Type 1. Journal of Virology,75(9), 4258-4267. doi:10.1128/jvi.75.9.4258-4267
Acknowledgements
I would like to thank Dr. Dahlquist for her help on this research project as well as Courtney Merriam's.While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.