Sbb14-Akash Jain
Tuesday Feb. 13
The bacteria chosen is: Thermus thermophilus
10 oligos pet team.
Each individual will be working on 2 oligos for two seperate Amino Acids.
I've been assigned Q (Glutamine) and Y (Tyrosine)
Thursday Feb. 15
We worked on finding the sequences that would encode those amino acids.
We found the complete genome for Thermus Thermophilus, found here: http://www.ncbi.nlm.nih.gov/nuccore/NC_006461.1?report=gbwithparts&log$=seqview&sat=4
Using the complete genome, we found sequences for the individual genes.
>The gene that encodes for the Glutamyl-tRNA synthetase: TTHA0438 atggtggtgacccgcatcgcgccaagccccacgggcgacccccacgtgggcacggcctacatcgccctcttcaactacgcctgggcccggaggaacgggg ggcgcttcatcgtgcgcattgaggacacggaccgggcgcgctacgtccccggggccgaggaaaggatcctcgccgccttaaagtggcttggcctctccta cgacgagggccccgatgtgggcggcccccacgggccctaccgccagtcggaacggcttcccctctaccaaaagtacgccgaggagctcctcaagcggggg tgggcctaccgggcctttgagaccccggaggagctagagcaaatccgcaaggaaaaggggggctacgacgggagggcccggaacatcccccctgaggagg ccgaggagcgggcgaggcggggcgagccccacgtgatccgcctcaaggtgccccgccccgggaccacggaggtcaaggacgagctcaggggggtggtggt ctacgacaaccaggagatccccgacgtggtcctcctcaagtccgacggctaccccacctaccacctggccaacgtggtggacgaccacctcatgggggtc acggacgtgatccgggcggaggagtggctcgtctccacccccatccacgtcctcctctaccgggccttcggctgggaggcgccccggttctaccacatgc ccctcctgcgcaaccccgacaagaccaagatctccaagcgcaaaagccacacctccctggactggtacaaggcggaggggtttctgcccgaggccctgag gaactacctctgcctcatggggttctccatgcccgacgggcgggagatcttcaccctcgaggagttcatccaggccttcacctgggagagggtttccctg ggggggcccgtctttgacctggaaaagctccgctggatgaacgggaagtacatccgggaggtgctctccctggaggaggtggcggagagggtcaagccct tcctccgggaggcggggctttcctgggaaagcgaggcctacctgaggcgggcggtggagctcatgcgcccccggtttgacaccctgaaggagttcccgga gaaggcccgctacctcttcaccgaggactaccccgtgtcggagaaggcccagaggaagctggaagaggggcttcccctcctcaaggagctctacccccgc ctaagggcccaggaggagtggaccgaggccgccctcgaggccctcctccggggcttcgccgcggagaagggggtcaagctcggccaggtggcccagcccc ttcgggccgccctcacggggagcctggagaccccggggctttttgagatcctggccctcctcgggaaggagcgggcgctcaggcgcttggagcgggccct cgcctag
>The gene that encodes for the Tyrosyl-tRNA synthetase: TTHA1399 atggcgggcaccggccacaccccggaagaggccctggccctcctcaagcggggggccgaggagatcgtccccgaggaagagcttctcgccaagctcaagg aggggcggcccctcacggtcaagctcggagccgaccccacgaggcccgacctgcacctgggccacgcggtggtcctgaggaagatgcgccagttccagga gctcggccacaaggtggtcctcatcatcggcgacttcaccgggatgatcggggacccttcgggccgttccaagacccggccccccctcaccctggaggag acccgggagaacgccaagacctacgtggcccaggcggggaagatcctcaggcaggagccccacctctttgagctccgctacaactccgagtggctggagg gcctcaccttcaaggaggtggtgcgcctcacctccctcatgaccgtggcccagatgctggaaagggaggacttcaagaagcggtacgaggcggggattcc catctccctgcacgagcttttgtaccccttcgcccaggcctacgactccgtggccataagggccgacgtggaaatggggggcacggaccagcgcttcaac ctcctggtgggccgggaggtgcaacgggcctacgggcaaagcccccaggtctgcttcctcatgccccttctcgtgggccttgacgggcgggagaagatga gcaagagcctggacaactacatcggcctcaccgagcccccggaggcgatgttcaagaagctcatgcgggtgccggaccctctcctcccgagctacttccg gcttctcacggacctggaggaggaggaaatagaggccctcctaaaggcgggccccgtccccgcccaccgggtcctcgcccgcctcctcaccgcggcctac gccctgccccagatccccccccggatagaccgggccttttacgaaagcctcggctacgcctgggaggccttcgggcgggacaaggaggcgggccccgagg aggtaaggagggcggaagcccgctacgacgaggtggccaaagggggaatccccgaggagatccccgaggtcaccatccccgcctcggagctgaaggaagg ccggatctgggtggcgaggctttttaccttagcgggcctcaccccctccaacgccgaggcgaggaggctcatccagaaccggggcctgaggctggacggg gaggtcctcaccgaccccatgctccaggtggacctctcccggccccgcatcctgcagcggggcaaggaccgcttcgtgcgggtgcggctttcggactga
Tuesday Feb. 20
We worked on finishing our oligos, created the forward and reverse models for our individual genes.
The sequences are as follows:
>Glutamine Forward gatctCCATGGtggtgacccgcatcgc
>Glutamine Reverse ctgatGAATTCctaggcgagggcccgctcca
>Tyrosine Forward catgtCCATGGcgggcaccggccacac
>Tyrosine Reverse gtcatGAATTCtcagtccgaaagccgcacc
We also proofread each other's to ensure that our oligos are all correct.
Thursday, Feb. 27
Watched demo of mini prep and gel purification. We did our own mini prep in the remaining time.
Tuesday, Mar. 4
Set up our first batch of PCR. Made the master mix, diluted our oligos by a factor of 100x. To do this, we diluted it 20 times initially, and then 5 times again. Once we created the PCR product, we put it in the thermocycler to perform the reaction.
Thursday, Mar. 6
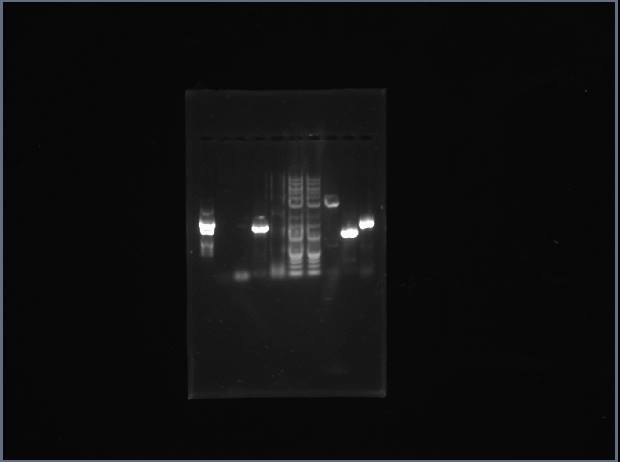
We ran a PCR for our group. Out of the syntases, approximately half failed. My tyrosine succeeded (last lane), but my guanine did not (2nd lane). I will have to reperform PCR on my product.
Friday, Mar. 7
Set up the PCR reactions for the failed synethetase for Guanine.
We also did Zymo Cleanup for our successful products, which for me was Tyrosine.
Tuesday, Mar. 11
Ran gel on the 2nd guanine PCR gel. This one also showed no good results. I'll have to try again.
Thursday, Mar. 13
Digested Tyrosine with EcoR1 and Nco1, and then did Zymo Cleanup of the digested product.
Tuesday, Mar. 18
Did a digestion of our previous mini prep, and incubated the product. All three wells were successful.
I set up another PCR reaction for guanine in the hopes that it will work this time. Had to remake "master mix."
Thursday, Mar. 20
We finished the Zymo Gel Purification on the wector digestion. I ligated Tyrosine with the vector digestion using ligaction process.
I set up the gel for the PCR. There are still no bands. I may have to try something new.
Thursday, Apr. 3
Plates did not turn out well. Redoing digestion.
PCR cannot be found. I'll have to run another PCR on my Q.
Friday, Apr. 4
Plates succeeded!
My Tyrosine infected E Coli are on the right most plate.
Unfortunately my PCR is gone yet again. Is someone stealing them? I've set up another PCR for the time being.
Tuesday, Apr. 8
I isolated the colonies for the tyrosine E. Coli into test tubes.
Ran the gel for the PCR. Did not turn out successful.
Thursday, Apr. 10
Miniprep on the colonies.
Tuesday, Apr. 15
Created a new PCR
Due to a shortage of gel boxes, we had to run zymo cleanup on our product.
Tursday, Apr. 17
Ran gel for PCR product and digested vector products.
The right most three are: Ysquare Ycircle PCRQ
SUCCESSFUL! The Q has actual product! I'm so happy.
Ycircle is also very good.
Friday, Apr. 18
After lecture, I performed Zymo cleanup on the Q from the PCR, and then subsequently the digestion.
Gel after digestion unfortunately did not turn out well. I will have to run digestion again before I can ligate.
Thursday, Apr. 25
Gave sample and sequence to Prof. Anderson for sequencing. Waiting on results.
>For the Y, the sequence should be: CATGGcgggcaccggccacaccccggaagaggccctggccctcctcaagcggggggccgaggagatcgtccccgaggaagagcttctcgccaagctcaag gaggggcggcccctcacggtcaagctcggagccgaccccacgaggcccgacctgcacctgggccacgcggtggtcctgaggaagatgcgccagttccagg agctcggccacaaggtggtcctcatcatcggcgacttcaccgggatgatcggggacccttcgggccgttccaagacccggccccccctcaccctggagga gacccgggagaacgccaagacctacgtggcccaggcggggaagatcctcaggcaggagccccacctctttgagctccgctacaactccgagtggctggag ggcctcaccttcaaggaggtggtgcgcctcacctccctcatgaccgtggcccagatgctggaaagggaggacttcaagaagcggtacgaggcggggattc ccatctccctgcacgagcttttgtaccccttcgcccaggcctacgactccgtggccataagggccgacgtggaaatggggggcacggaccagcgcttcaa cctcctggtgggccgggaggtgcaacgggcctacgggcaaagcccccaggtctgcttcctcatgccccttctcgtgggccttgacgggcgggagaagatg agcaagagcctggacaactacatcggcctcaccgagcccccggaggcgatgttcaagaagctcatgcgggtgccggaccctctcctcccgagctacttcc ggcttctcacggacctggaggaggaggaaatagaggccctcctaaaggcgggccccgtccccgcccaccgggtcctcgcccgcctcctcaccgcggccta cgccctgccccagatccccccccggatagaccgggccttttacgaaagcctcggctacgcctgggaggccttcgggcgggacaaggaggcgggccccgag gaggtaaggagggcggaagcccgctacgacgaggtggccaaagggggaatccccgaggagatccccgaggtcaccatccccgcctcggagctgaaggaag gccggatctgggtggcgaggctttttaccttagcgggcctcaccccctccaacgccgaggcgaggaggctcatccagaaccggggcctgaggctggacgg ggaggtcctcaccgaccccatgctccaggtggacctctcccggccccgcatcctgcagcggggcaaggaccgcttcgtgcgggtgcggctttcggactga GAATTCGAAGCTTGGGCCCGAACAAAAACTCATCTCAGAAGAGGATCTGAATAGCGCCGTCGACCATCATCATCATCATCATTGAGTTTAAACGGTCTCC AGCTTGGCTGTTTTGGCGGATGAGAGAAGATTTTCAGCCTGATACAGATTAAATCAGAACGCAGAAGCGGTCTGATAAAACAGAATTTGCCTGGCGGCAG TAGCGCGGTGGTCCCACCTGACCCCATGCCGAACTCAGAAGTGAAACGCCGTAGCGCCGATGGTAGTGTGGGGTCTCCCCATGCGAGAGTAGGGAACTGC CAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCCTGAGTAGGACAAATCCGCCG GGAGCGGATTTGAACGTTGCGAAGCAACGGCCCGGAGGGTGGCGGGCAGGACGCCCGCCATAAACTGCCAGGCATCAAATTAAGCAGAAGGCCATCCTGA CGGATGGCCTTTTTGCGTTTCTACAAACTCTTTTTGTTTATTTTTCTAAATACATTCAAATATGTATCCGCTCATGAGACAATAACCCTGATAAATGCTT CAATAATATTGAAAAAGGAAGAGTATGAGTATTCAACATTTCCGTGTCGCCCTTATTCCCTTTTTTGCGGCATTTTGCCTTCCTGTTTTTGCTCACCCAG AAACGCTGGTGAAAGTAAAAGATGCTGAAGATCAGTTGGGTGCACGAGTGGGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAGAGTTTTCG CCCCGAAGAACGTTTTCCAATGATGAGCACTTTTAAAGTTCTGCTATGTGGCGCGGTATTATCCCGTGTTGACGCCGGGCAAGAGCAACTCGGTCGCCGC ATACACTATTCTCAGAATGACTTGGTTGAGTACTCACCAGTCACAGAAAAGCATCTTACGGATGGCATGACAGTAAGAGAATTATGCAGTGCTGCCATAA CCATGAGTGATAACACTGCGGCCAACTTACTTCTGACAACGATCGGAGGACCGAAGGAGCTAACCGCTTTTTTGCACAACATGGGGGATCATGTAACTCG CCTTGATCGTTGGGAACCGGAGCTGAATGAAGCCATACCAAACGACGAGCGTGACACCACGATGCCTGTAGCAATGGCAACAACGTTGCGCAAACTATTA ACTGGCGAACTACTTACTCTAGCTTCCCGGCAACAATTAATAGACTGGATGGAGGCGGATAAAGTTGCAGGACCACTTCTGCGCTCGGCCCTTCCGGCTG GCTGGTTTATTGCTGATAAATCTGGAGCCGGTGAGCGTGGGTCTCGCGGTATCATTGCAGCACTGGGGCCAGATGGTAAGCCCTCCCGTATCGTAGTTAT CTACACGACGGGGAGTCAGGCAACTATGGATGAACGAAATAGACAGATCGCTGAGATAGGTGCCTCACTGATTAAGCATTGGTAACTGTCAGACCAAGTT TACTCATATATACTTTAGATTGATTTAAAACTTCATTTTTAATTTAAAAGGATCTAGGTGAAGATCCTTTTTGATAATCTCATGACCAAAATCCCTTAAC GTGAGTTTTCGTTCCACTGAGCGTCAGACCCCGTAGAAAAGATCAAAGGATCTTCTTGAGATCCTTTTTTTCTGCGCGTAATCTGCTGCTTGCAAACAAA AAAACCACCGCTACCAGCGGTGGTTTGTTTGCCGGATCAAGAGCTACCAACTCTTTTTCCGAAGGTAACTGGCTTCAGCAGAGCGCAGATACCAAATACT GTCCTTCTAGTGTAGCCGTAGTTAGGCCACCACTTCAAGAACTCTGTAGCACCGCCTACATACCTCGCTCTGCTAATCCTGTTACCAGTGGCTGCTGCCA GTGGCGATAAGTCGTGTCTTACCGGGTTGGACTCAAGACGATAGTTACCGGATAAGGCGCAGCGGTCGGGCTGAACGGGGGGTTCGTGCACACAGCCCAG CTTGGAGCGAACGACCTACACCGAACTGAGATACCTACAGCGTGAGCTATGAGAAAGCGCCACGCTTCCCGAAGGGAGAAAGGCGGACAGGTATCCGGTA AGCGGCAGGGTCGGAACAGGAGAGCGCACGAGGGAGCTTCCAGGGGGAAACGCCTGGTATCTTTATAGTCCTGTCGGGTTTCGCCACCTCTGACTTGAGC GTCGATTTTTGTGATGCTCGTCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCTGGCCTTTTGCTCA CATGTTCTTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCCTTTGAGTGAGCTGATACCGCTCGCCGCAGCCGAACGACCGAGCGCAGC GAGTCAGTGAGCGAGGAAGCGGAAGAGCGCCTGATGCGGTATTTTCTCCTTACGCATCTGTGCGGTATTTCACACCGCATATGGTGCACTCTCAGTACAA TCTGCTCTGATGCCGCATAGTTAAGCCAGTATACACTCCGCTATCGCTACGTGACTGGGTCATGGCTGCGCCCCGACACCCGCCAACACCCGCTGACGCG CCCTGACGGGCTTGTCTGCTCCCGGCATCCGCTTACAGACAAGCTGTGACCGTCTCCGGGAGCTGCATGTGTCAGAGGTTTTCACCGTCATCACCGAAAC GCGCGAGGCAGCAGATCAATTCGCGCGCGAAGGCGAAGCGGCATGCATAATGTGCCTGTCAAATGGACGAAGCAGGGATTCTGCAAACCCTATGCTACTC CGTCAAGCCGTCAATTGTCTGATTCGTTACCAATTATGACAACTTGACGGCTACATCATTCACTTTTTCTTCACAACCGGCACGGAACTCGCTCGGGCTG GCCCCGGTGCATTTTTTAAATACCCGCGAGAAATAGAGTTGATCGTCAAAACCAACATTGCGACCGACGGTGGCGATAGGCATCCGGGTGGTGCTCAAAA GCAGCTTCGCCTGGCTGATACGTTGGTCCTCGCGCCAGCTTAAGACGCTAATCCCTAACTGCTGGCGGAAAAGATGTGACAGACGCGACGGCGACAAGCA AACATGCTGTGCGACGCTGGCGATATCAAAATTGCTGTCTGCCAGGTGATCGCTGATGTACTGACAAGCCTCGCGTACCCGATTATCCATCGGTGGATGG AGCGACTCGTTAATCGCTTCCATGCGCCGCAGTAACAATTGCTCAAGCAGATTTATCGCCAGCAGCTCCGAATAGCGCCCTTCCCCTTGCCCGGCGTTAA TGATTTGCCCAAACAGGTCGCTGAAATGCGGCTGGTGCGCTTCATCCGGGCGAAAGAACCCCGTATTGGCAAATATTGACGGCCAGTTAAGCCATTCATG CCAGTAGGCGCGCGGACGAAAGTAAACCCACTGGTGATACCATTCGCGAGCCTCCGGATGACGACCGTAGTGATGAATCTCTCCTGGCGGGAACAGCAAA ATATCACCCGGTCGGCAAACAAATTCTCGTCCCTGATTTTTCACCACCCCCTGACCGCGAATGGTGAGATTGAGAATATAACCTTTCATTCCCAGCGGTC GGTCGATAAAAAAATCGAGATAACCGTTGGCCTCAATCGGCGTTAAACCCGCCACCAGATGGGCATTAAACGAGTATCCCGGCAGCAGGGGATCATTTTG CGCTTCAGCCATACTTTTCATACTCCCGCCATTCAGAGAAGAAACCAATTGTCCATATTGCATCAGACATTGCCGTCACTGCGTCTTTTACTGGCTCTTC TCGCTAACCAAACCGGTAACCCCGCTTATTAAAAGCATTCTGTAACAAAGCGGGACCAAAGCCATGACAAAAACGCGTAACAAAAGTGTCTATAATCACG GCAGAAAAGTCCACATTGATTATTTGCACGGCGTCACACTTTGCTATGCCATAGCATTTTTATCCATAAGATTAGCGGATCCTACCTGACGCTTTTTATC GCAACTCTCTACTGTTTCTCCATACCCGTTTTTTTGGGCTAACAGGAGGAATTAAC
For the Q, used the eluted product to run digestion. Ran ligation on the digestion product, and did transformation subsequently. Out of plates, so I will plate tomorrow.
Friday, Apr. 27
Plate had colonies. I will pick these out and incubate in LB media with antibioitic till next week.
Tuesday, Apr. 29
Recieved my sequence back from the Y sample and used [[Georgetown's Pairwise Alignment Tool[1]]] to compare the sequences. It found a 99.9% of match for a region of 836 bp (with one minor misread downstream).
Results:
Picked 4 colonies of the Q and put them for incubation in 5 ml of media. I will run the miniprep purification and digestion on the product on thursday, and submit my product.
>Expected DNA for the Q: CATGGtggtgacccgcatcgcgccaagccccacgggcgacccccacgtgggcacggcctacatcgccctcttcaactacgcctgggcccggaggaacggg gggcgcttcatcgtgcgcattgaggacacggaccgggcgcgctacgtccccggggccgaggaaaggatcctcgccgccttaaagtggcttggcctctcct acgacgagggccccgatgtgggcggcccccacgggccctaccgccagtcggaacggcttcccctctaccaaaagtacgccgaggagctcctcaagcgggg gtgggcctaccgggcctttgagaccccggaggagctagagcaaatccgcaaggaaaaggggggctacgacgggagggcccggaacatcccccctgaggag gccgaggagcgggcgaggcggggcgagccccacgtgatccgcctcaaggtgccccgccccgggaccacggaggtcaaggacgagctcaggggggtggtgg tctacgacaaccaggagatccccgacgtggtcctcctcaagtccgacggctaccccacctaccacctggccaacgtggtggacgaccacctcatgggggt cacggacgtgatccgggcggaggagtggctcgtctccacccccatccacgtcctcctctaccgggccttcggctgggaggcgccccggttctaccacatg cccctcctgcgcaaccccgacaagaccaagatctccaagcgcaaaagccacacctccctggactggtacaaggcggaggggtttctgcccgaggccctga ggaactacctctgcctcatggggttctccatgcccgacgggcgggagatcttcaccctcgaggagttcatccaggccttcacctgggagagggtttccct gggggggcccgtctttgacctggaaaagctccgctggatgaacgggaagtacatccgggaggtgctctccctggaggaggtggcggagagggtcaagccc ttcctccgggaggcggggctttcctgggaaagcgaggcctacctgaggcgggcggtggagctcatgcgcccccggtttgacaccctgaaggagttcccgg agaaggcccgctacctcttcaccgaggactaccccgtgtcggagaaggcccagaggaagctggaagaggggcttcccctcctcaaggagctctacccccg cctaagggcccaggaggagtggaccgaggccgccctcgaggccctcctccggggcttcgccgcggagaagggggtcaagctcggccaggtggcccagccc cttcgggccgccctcacggggagcctggagaccccggggctttttgagatcctggccctcctcgggaaggagcgggcgctcaggcgcttggagcgggccc tcgcctagGAATTCGAAGCTTGGGCCCGAACAAAAACTCATCTCAGAAGAGGATCTGAATAGCGCCGTCGACCATCATCATCATCATCATTGAGTTTAAA CGGTCTCCAGCTTGGCTGTTTTGGCGGATGAGAGAAGATTTTCAGCCTGATACAGATTAAATCAGAACGCAGAAGCGGTCTGATAAAACAGAATTTGCCT GGCGGCAGTAGCGCGGTGGTCCCACCTGACCCCATGCCGAACTCAGAAGTGAAACGCCGTAGCGCCGATGGTAGTGTGGGGTCTCCCCATGCGAGAGTAG GGAACTGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCCTGAGTAGGACAA ATCCGCCGGGAGCGGATTTGAACGTTGCGAAGCAACGGCCCGGAGGGTGGCGGGCAGGACGCCCGCCATAAACTGCCAGGCATCAAATTAAGCAGAAGGC CATCCTGACGGATGGCCTTTTTGCGTTTCTACAAACTCTTTTTGTTTATTTTTCTAAATACATTCAAATATGTATCCGCTCATGAGACAATAACCCTGAT AAATGCTTCAATAATATTGAAAAAGGAAGAGTATGAGTATTCAACATTTCCGTGTCGCCCTTATTCCCTTTTTTGCGGCATTTTGCCTTCCTGTTTTTGC TCACCCAGAAACGCTGGTGAAAGTAAAAGATGCTGAAGATCAGTTGGGTGCACGAGTGGGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAG AGTTTTCGCCCCGAAGAACGTTTTCCAATGATGAGCACTTTTAAAGTTCTGCTATGTGGCGCGGTATTATCCCGTGTTGACGCCGGGCAAGAGCAACTCG GTCGCCGCATACACTATTCTCAGAATGACTTGGTTGAGTACTCACCAGTCACAGAAAAGCATCTTACGGATGGCATGACAGTAAGAGAATTATGCAGTGC TGCCATAACCATGAGTGATAACACTGCGGCCAACTTACTTCTGACAACGATCGGAGGACCGAAGGAGCTAACCGCTTTTTTGCACAACATGGGGGATCAT GTAACTCGCCTTGATCGTTGGGAACCGGAGCTGAATGAAGCCATACCAAACGACGAGCGTGACACCACGATGCCTGTAGCAATGGCAACAACGTTGCGCA AACTATTAACTGGCGAACTACTTACTCTAGCTTCCCGGCAACAATTAATAGACTGGATGGAGGCGGATAAAGTTGCAGGACCACTTCTGCGCTCGGCCCT TCCGGCTGGCTGGTTTATTGCTGATAAATCTGGAGCCGGTGAGCGTGGGTCTCGCGGTATCATTGCAGCACTGGGGCCAGATGGTAAGCCCTCCCGTATC GTAGTTATCTACACGACGGGGAGTCAGGCAACTATGGATGAACGAAATAGACAGATCGCTGAGATAGGTGCCTCACTGATTAAGCATTGGTAACTGTCAG ACCAAGTTTACTCATATATACTTTAGATTGATTTAAAACTTCATTTTTAATTTAAAAGGATCTAGGTGAAGATCCTTTTTGATAATCTCATGACCAAAAT CCCTTAACGTGAGTTTTCGTTCCACTGAGCGTCAGACCCCGTAGAAAAGATCAAAGGATCTTCTTGAGATCCTTTTTTTCTGCGCGTAATCTGCTGCTTG CAAACAAAAAAACCACCGCTACCAGCGGTGGTTTGTTTGCCGGATCAAGAGCTACCAACTCTTTTTCCGAAGGTAACTGGCTTCAGCAGAGCGCAGATAC CAAATACTGTCCTTCTAGTGTAGCCGTAGTTAGGCCACCACTTCAAGAACTCTGTAGCACCGCCTACATACCTCGCTCTGCTAATCCTGTTACCAGTGGC TGCTGCCAGTGGCGATAAGTCGTGTCTTACCGGGTTGGACTCAAGACGATAGTTACCGGATAAGGCGCAGCGGTCGGGCTGAACGGGGGGTTCGTGCACA CAGCCCAGCTTGGAGCGAACGACCTACACCGAACTGAGATACCTACAGCGTGAGCTATGAGAAAGCGCCACGCTTCCCGAAGGGAGAAAGGCGGACAGGT ATCCGGTAAGCGGCAGGGTCGGAACAGGAGAGCGCACGAGGGAGCTTCCAGGGGGAAACGCCTGGTATCTTTATAGTCCTGTCGGGTTTCGCCACCTCTG ACTTGAGCGTCGATTTTTGTGATGCTCGTCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCTGGCCT TTTGCTCACATGTTCTTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCCTTTGAGTGAGCTGATACCGCTCGCCGCAGCCGAACGACCG AGCGCAGCGAGTCAGTGAGCGAGGAAGCGGAAGAGCGCCTGATGCGGTATTTTCTCCTTACGCATCTGTGCGGTATTTCACACCGCATATGGTGCACTCT CAGTACAATCTGCTCTGATGCCGCATAGTTAAGCCAGTATACACTCCGCTATCGCTACGTGACTGGGTCATGGCTGCGCCCCGACACCCGCCAACACCCG CTGACGCGCCCTGACGGGCTTGTCTGCTCCCGGCATCCGCTTACAGACAAGCTGTGACCGTCTCCGGGAGCTGCATGTGTCAGAGGTTTTCACCGTCATC ACCGAAACGCGCGAGGCAGCAGATCAATTCGCGCGCGAAGGCGAAGCGGCATGCATAATGTGCCTGTCAAATGGACGAAGCAGGGATTCTGCAAACCCTA TGCTACTCCGTCAAGCCGTCAATTGTCTGATTCGTTACCAATTATGACAACTTGACGGCTACATCATTCACTTTTTCTTCACAACCGGCACGGAACTCGC TCGGGCTGGCCCCGGTGCATTTTTTAAATACCCGCGAGAAATAGAGTTGATCGTCAAAACCAACATTGCGACCGACGGTGGCGATAGGCATCCGGGTGGT GCTCAAAAGCAGCTTCGCCTGGCTGATACGTTGGTCCTCGCGCCAGCTTAAGACGCTAATCCCTAACTGCTGGCGGAAAAGATGTGACAGACGCGACGGC GACAAGCAAACATGCTGTGCGACGCTGGCGATATCAAAATTGCTGTCTGCCAGGTGATCGCTGATGTACTGACAAGCCTCGCGTACCCGATTATCCATCG GTGGATGGAGCGACTCGTTAATCGCTTCCATGCGCCGCAGTAACAATTGCTCAAGCAGATTTATCGCCAGCAGCTCCGAATAGCGCCCTTCCCCTTGCCC GGCGTTAATGATTTGCCCAAACAGGTCGCTGAAATGCGGCTGGTGCGCTTCATCCGGGCGAAAGAACCCCGTATTGGCAAATATTGACGGCCAGTTAAGC CATTCATGCCAGTAGGCGCGCGGACGAAAGTAAACCCACTGGTGATACCATTCGCGAGCCTCCGGATGACGACCGTAGTGATGAATCTCTCCTGGCGGGA ACAGCAAAATATCACCCGGTCGGCAAACAAATTCTCGTCCCTGATTTTTCACCACCCCCTGACCGCGAATGGTGAGATTGAGAATATAACCTTTCATTCC CAGCGGTCGGTCGATAAAAAAATCGAGATAACCGTTGGCCTCAATCGGCGTTAAACCCGCCACCAGATGGGCATTAAACGAGTATCCCGGCAGCAGGGGA TCATTTTGCGCTTCAGCCATACTTTTCATACTCCCGCCATTCAGAGAAGAAACCAATTGTCCATATTGCATCAGACATTGCCGTCACTGCGTCTTTTACT GGCTCTTCTCGCTAACCAAACCGGTAACCCCGCTTATTAAAAGCATTCTGTAACAAAGCGGGACCAAAGCCATGACAAAAACGCGTAACAAAAGTGTCTA TAATCACGGCAGAAAAGTCCACATTGATTATTTGCACGGCGTCACACTTTGCTATGCCATAGCATTTTTATCCATAAGATTAGCGGATCCTACCTGACGC TTTTTATCGCAACTCTCTACTGTTTCTCCATACCCGTTTTTTTGGGCTAACAGGAGGAATTAAC
Thursday, May 1
Did miniprep and digestion of 4 colonies, and created the final products.
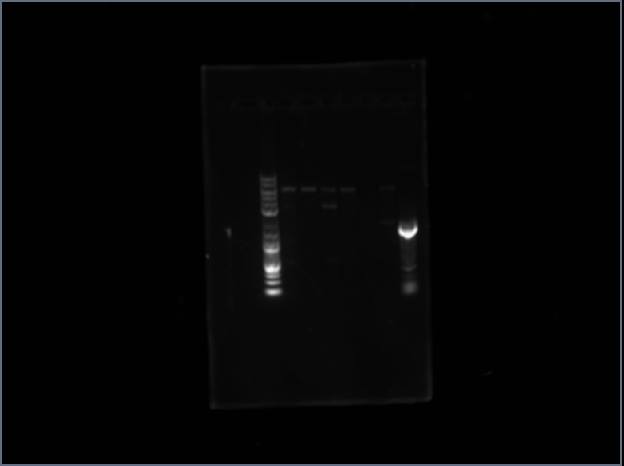
I ran a gel, here are the results:
Qa Qb Ladder Qx Qy
It's faint, but Qx and Qy are present.
I submitted Qx to Chris for sequencing.
Friday, May 2
Recieved my sequence back from the Q sample and used [[Georgetown's Pairwise Alignment Tool[2]]] to compare the sequences. It found a 97.6% of match for a region of 164 bp.
Results: