SMGT-ODE: the full text
DNA interaction with sperm cells: ODE model
Author: Andrew Kuznetsov
Affiliation: Institute of Biology III, Albert-Ludwig’s University Freiburg,
Schaenzlestrasse 1, D-79104 Freiburg, Germany
Contact email: andrei.kouznetsov@imtek.uni-freiburg.de
Keywords: sperm mediated gene transfer, ODE model, compositional evolution
Abstract
A computer simulation concerning initial events in the sperm/DNA interaction was performed. DNA binding with MHCII and CD4 proteins, DNA internalization into spermatozoa, and DNase activity in sperm were described by first order differential equations (ODE). The dynamics of the system depended on the parameters of the model. The simulation could explain controversial results in sperm mediated gene transfer research and posed this phenomenon to an unknown form of biological evolution.
Introduction
The phenomenon of foreign gene transfer with sperm was first observed in lab conditions in 1971 [1]. Since 1989 [2, 3], when sperm mediated gene transfer (SMGT) was rediscovered, some unusual molecular mechanisms in spermatozoa were described, such as the ability of sperm cells to capture exogenous DNA [4] and RNA [5] molecules, the reverse transcriptase activity in sperm cells [6], the insertion of foreign nucleic acids into sperm chromatin [7] and an apoptosis-like response to the high concentration of internalized DNA [8]. SMGT has been seen as a mechanism for genome destabilization [9].
The reason of a particular interest to this topic is the obvious possibility to create a simple method for mass transgenesis. The number of publications about SMGT is increasing [10]. However, the theoretical interest to SMGT in scientific community grows slowly because of paradigm that “SMGT can lead to a chaos in the biological world”. From another point of view a non-linear dynamics is the main feature of biological systems, which leads to a self-organization far from the equilibrium state [11]. In this context we considered SMGT as an example of the process concerning loss of stability, chaos and evolution [12]. Even an agent based toy model of spermatozoa and ova crossing with ‘modulo mutations’ demonstrated the non-trivial behavior–periodic and chaotic variations for an average digital genome [13].
Molecular mechanisms of sperm/DNA interaction are partly known (Fig. 1a). Originally, glycoprotein Inhibitor Factor I (IF-1) competes for DNA binding sites on a sperm surface and prevents DNA interaction with sperm cells [14]. In the absence of seminal fluid that contains IF-1 protein and nucleases [15], foreign DNA binds strongly (Kd=7×10-15 M) to major histocompatibility complex class II molecules (MHCII), which are exposed on the sperm membrane [16]. DNA interaction with CD4 receptor leads to internalization of the protein/DNA complex into sperm cells [17], where DNA can be attacked by internal nucleases [6]. When foreign DNA penetrates into a sperm nucleus [18] it can attain an episomal form [19] or may be inserted into the sperm chromatin [8]. Fate of foreign DNA during embryogenesis and its transfer into progeny are of great interest, but this is not a subject of this report. Here, I suggest an initial sperm/DNA interaction scenario that might partially explain the controversial results of SMGT efficiency, which varied in some experiments from 0 to more than 85% [20]. In this work the dynamics of sperm/DNA interaction was investigated by computer simulation.
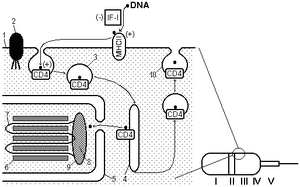

Fig. 1 A–Hypothetical mechanism for DNA penetration into spermatozoon [21]. 1 - plasmalemma, 2 - cytoskeleton binding protein, 3 - vesicle, 4 - releasing of DNA from vesicle into cytoplasm, 5 - nuclear envelope, 6 - protamines, 7 - chromosomal DNA loop, 8 - scaffold, 9 - nuclear annulus, 10 - releasing of DNA from vesicle into environment; +/– - increase / decrease of DNA uptake. Parts of spermatozoon: I - acrosomal cap, II - equatorial segment, III - postacrosomal region, IV - posterior ring, V - middle piece. B–Flow diagram of proteins/DNA interactions in sperm cell.
Method
The structural model of sperm/DNA interaction (Fig. 1a) was used to describe the problem. I assumed free foreign DNA, complexes of DNA with MHCII and CD4 proteins, DNA compartmentalization in the sperm cell, DNase activities in the seminal fluid and in the spermatozoon. The DNA flow between nodes, which are represented on the graph in Fig. 1b, was described by the first order differential equations:
dS1/dt = k-1*S2 - (k1 + k5 + k12)*S1 + k4*S4
dS2/dt = k1*S1 - (k-1 + k2)*S2
dS3/dt = k12*S1 + k2*S2 - k3*S3
dS4/dt = k3*S3 - (k4 + k6)*S4
dS5/dt = k5*S1
dS6/dt = k6*S4 (1)
where, variable S1 is free DNA, S2 is MHCII bound DNA, S3 is CD4 bound DNA, S4 is foreign DNA in the sperm cell, variables S5 and S6 are DNAs cleaved by DNases outside and inside sperm cells respectively. Ratios of parameters k-1 = k4 = 0.1 were not varied; values of parameters k5 and k6 were changed during ‘physiological’ experiments with DNase activities: k5 = {0, 1}, k6 = {0, 0.1, 1}; the coefficients k1, k2, k12, k3 were changed in ‘genetic’ experiments and in the simulation of IF-1 block: k1 = {0, 1}, k2 = k12 = k3 = {0, 0.1}. The initial conditions were 1 for input DNA (S1 = 1) and 0 for all DNA binding contents, {S2,...,S6} = 0. Combinations of parameter values were investigated by the simulation.
To resolve the system of differential equations (1) the numerical Runge-Kutta method was used [22]. The simulation and data representation were implemented in Matlab7 environment. Time intervals (t) were denoted by calculation steps of SMGT1 program (Appendix), the concentrations were represented in arbitrary units (c) on the range [0, 1]. Time in silico corresponded to time of reaction in vitro was estimated in minutes; virtual concentrations accompanied portions of added DNA in real biological experiments.
Results
The kinetic of DNA binding by sperm cells was analyzed at k1 = 1, k-1 = k2 = k12 = k3 = k4 = 0.1, k5 = k6 = 0 that is corresponded to the ideal laboratory conditions for SMGT, where was neither IF-1 nor any DNase activity in sperm. Total amount of free DNA decreased sharply to 0.07 c during the first 5 time intervals (t) because of interaction of DNA with sperm structures (Fig. 2, red line). I observed very fast binding of DNA to MHCII molecules spiked on 0.66 c at 2.5 t (blue line). The redistribution of bound DNA during DNA interaction with CD4 receptors was slow (max 0.39 c at 12 t, green line). A delay (lag phase) of DNA penetration into sperm cells was revealed as a result of early DNA binding to MHCII and CD4 receptors (about 3 t). The content of internalized DNA peaked on 0.33 c at 30 t (dashed magenta line). At this point the system remained steady. More then 90 % of total DNA was bound to MHCII, CD4, and within the internal section of sperm cells. The DNA was evenly distributed between these parts (about 30% for each one). The content of DNA linked with CD4 and the internal compartment of sperm cells (N.B., except to MHCII) was 0.38 + 0.26 = 0.64 c at 15 t. It confirms our experimental data when approximately 60% of externally added DNA was bound by rabbit sperm [23]. Thus, in a virtual experiment, the reaction leads to an increasing amount of foreign DNA in the sperm cells during first 30 time units until a plateau.
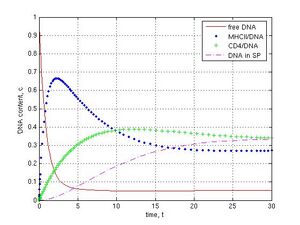
Fig. 2 DNA interaction with ‘washed’ sperm cells at following conditions: k1 =1, k-1 = k2 = k12 = k3 = k4 = 0.1, k5 = k6 = 0, reaction during 30 t. Concentration (c) and time (t) units are arbitrary. DNA binds very fast to MHCII proteins at the beginning of reaction, interacts more slowly with CD4 receptors, penetrates into sperm cell with a delay, and reaches the plateau in 30th time interval.
Then I simulated nucleases activities in seminal fluid. If the activity was high (k5 = 1) the system behaved in another way. Free DNA was declined to 0.004 c in presence of DNase activity as compared with DNA in absence of nucleases (0.06 c) (Fig. 3a and b, red lines). The maximal amount of DNA/MHCII complexes was decreased from 0.66 to 0.38 c (blue point lines), DNA/CD4 complexes–from 0.39 to 0.17 c (green plus lines), and DNA in the internal part of sperm cells–from 0.33 to 0.12 c (magenta dashed lines). I revealed that DNase activity in a virtual seminal fluid did not fully prevent DNA penetration into sperm cells and a small part of DNA reached the internal compartment of spermatozoa in first 20 t. Finally this DNA was slowly degraded because of its release from cells into external environment, where DNase activity was modeled.
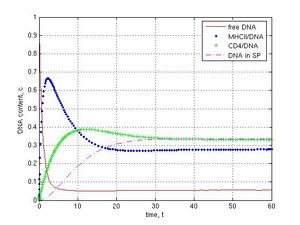
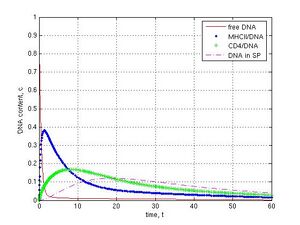
Fig. 3 Effect of DNases in seminal fluid on sperm/DNA interaction: A–in absence of DNase activity, k5 = 0; B–in presence of DNase activity in seminal fluid, k5 = 1. Other parameters were k1 = 1, k-1 = k2 = k12 = k3 = k4 = 0.1, k6 = 0. Unlike control A model B (k5 = 1) demonstrates fast degradation of foreign DNA.
DNase activity inside sperm cells was investigated at k1 = 1, k-1 = k12 = k2 = k3 = k4 = 0.1, k5 = 0. Different values of k6 were used for simulation. Absence of nucleases corresponded to k6 = 0, intermediate level of DNase activity was at k6 = 0.1 and high DNase activity was at k6 = 1. As it was shown, in the absence of DNase activity system reaches steady state in 30th time interval (see Fig. 2 and 3a). When intermediate DNase activity (k6 = 0.1) was simulated, the amount of internalized DNA decreased gradually. Content of DNA inside sperm cells came down from 0.17 at 18 t to 0.07 c at 60 t (Fig. 4a, magenta dashed line). In case of strong DNase activity (k6 = 1) only low of DNA (0.03 c at 11 t and less then 0.01 c at 60 t) were found in the internal part of spermatozoa (Fig. 4b, magenta dashed line).
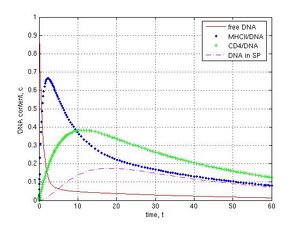
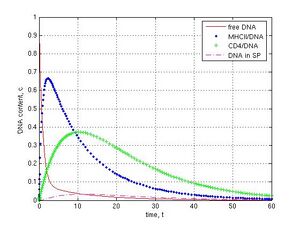
Fig. 4 Impact of internal DNases on the foreign DNA content in sperm cells (modeling at k1 = 1, k-1 = k12 = k2 = k3 = k4 = 0.1, k5 = 0): A–the slight decrease of the DNA amount at low DNase activity in sperm cells, k6 = 0.1; B–high DNase activity leads to drop in introduced DNA, k6 = 1. The variation in DNase activity (k6 value) provokes the different final DNA content in sperm cells (magenta dashed line).
To describe the dynamics of DNA interaction with MHCII and CD4 proteins, so as DNA internalization into spermatozoa, and it slow degradation by internal DNases the output was represented by a phase portrait, where immediate MHCII/DNA, CD4/DNA, and SP/DNA values were drawn on 3D graphic. Thus, a line in the 3D space (S2, S3, S4) described a behavior of the system (Fig. 5). The parameters were chosen like in the previous case: k1 = 1, k-1 = k12 = k2 = k3 = k4 = 0.1, k5 = 0, k6 = 0.1 (Fig. 4a).
I revealed three different stages in the sperm/DNA interaction traveling along the curve. At the first stage foreign DNA was bound very fast to MHCII. Curve moved along the x axis and reached the maximal MHCII/DNA value 0.65 c (0.65, 0.15, 0.01). At the second stage the curve turned into the y and z axis and took the point (0.32, 0.35, 0.15) at 15 t that is corresponded to maximum of SP/DNA content (black square). It proves that DNA interacts with CD4 receptor and then internalizes into sperm cell. At the third stage the line turned back to a start position along the x, y, z axis and arrived at the point (0.08, 0.13, 0.07) at 60th time interval. The line came to final point (0.02, 0.03, 0.02) at 120 t. A prolonged time of reaction led to a nearly total degradation of DNA because of DNase activity. On the other hand, if DNase activity was switched off (k6 = 0) the maximal amount of foreign DNA in sperm cells was 0.33 c with x, y coordinates (0.28, 0.33) after 60 t of simulation (like on Fig. 3a).
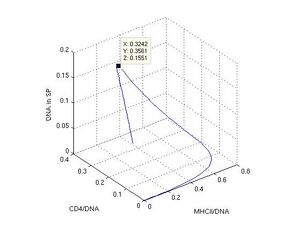
Fig. 5 3D phase diagram of bound DNA in coordinates (x, y, z), where x is MHCII, y is CD4, and z is the internal part of sperm cell. Values of MHCII/DNA, CD4/DNA, and SP/DNA are on the x, y, and z axis.
When knockout genetic experiments or IF-1 block in the ‘electronic’ sperm cell (Fig. 1b) were carried out at k-1 = k4 = 0.1, k5 = k6 = 0, the subsequent DNA transfer pathways were switched off by the assigning of zero values for corresponding coefficients. As shown, the block for DNA binding to MHCII (k1 = 0) did not prevent DNA penetration into sperm cells and led to the steady value 0.33 c, because of the parallel pathway for CD4 receptor (Fig. 1b and 6a, CD4–green pluses, SP–magenta dashed line). If MHCII interaction with CD4 was prevented (k2 = 0), DNA molecules were deposed on MHCII (0.77 c) and only a small part of DNA was found (0.08 c) in sperm cells after 30 t (Fig. 6b, blue point and magenta dashed lines). The inhibition of DNA binding to CD4 was achieved with the coefficients k12 = k2 = 0. In this case a fast exponential crash of free DNA to 0.09 c and a fast logarithmic growth of MHCII/DNA complexes to 0.90 c during 5 t were observed (Fig. 6c, red and blue point lines). If I interrupted the DNA internalization into spermatozoa (k3 = 0), DNA was deposed on CD4 receptors during 60 t (Fig. 6d, green plus line).
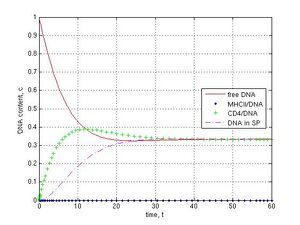
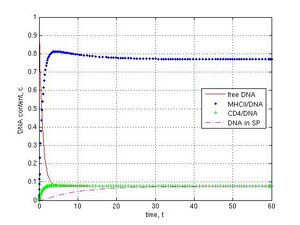
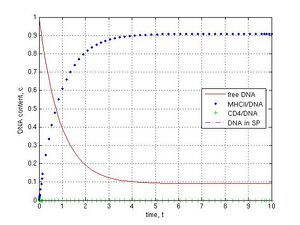
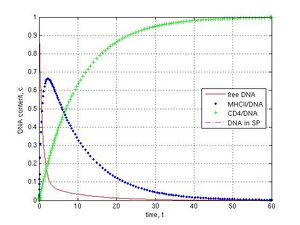
Fig. 6 Disruption of DNA transfer pathways (k-1 = k4 = 0.1, k5 = k6 = 0): A–MHCII/DNA block (k1 = 0), B–MHCII/CD4 block (k2 = 0), C–CD4/DNA block (k12 = k2 = 0), D–SP/DNA block (k3 = 0). No DNA in the internal part of sperm cells in C and D cases (no magenta dashed line).
Finally I investigated some imaginary in vivo scenarios. When I modeled the presence of IF-1 protein and a powerful activity of nucleases in the seminal fluid at k1= k12 = 0 and k5 = 1, which simulated protected conditions, a very fast exponential degradation of free DNA during 5 t and no foreign DNA in sperm cells were observed (Fig. 7a, red line). When, for example, the ratio for all parameters in the model was 0.1, which corresponds to ‘leaky’ conditions, the DNA content in the internal sperm compartment reached 0.12 c at 14 t that might led to a genetic transformation of sperm.
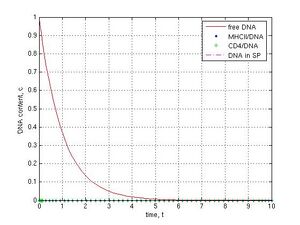
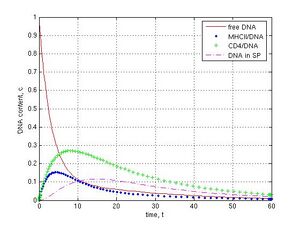
Fig. 7 Different SMGT scenarios: A–protected conditions (there are IF-1 and nucleases in seminal fluid: k1 = k12 = 0, k5 = 1), other coefficients were chosen k-1 = k2 = k3 = k4 = 0.1, k6 = 0; B–an unusual ‘open’ combination of parameters (k1 = ... = k6 = 0.1), which leads to the DNA penetration into sperm cells and a possible genetic transformation.
Discussion
I put some assumptions, based on experimental data, into the SMGT1 model, such as: 1) DNA binds to MHCII and CD4 molecules, 2) the CD4 receptor takes part in the DNA internalization into sperm cells, 3) IF-1 prevents DNA binding to MHCII and CD4, 4) there are DNase activities inside and outside sperm cells. I speculated about an interaction between MHCII and CD4 molecules and about the release of a part of the DNA from spermatozoa by CD4 recycling (Fig. 1a). However, the model of foreign DNA stream in spermatozoa (Fig. 1b) was in good agreement with experiments for sperm/DNA interaction [4, 17].
The modeling described some stages of DNA penetration into a sperm cell: delay phase, DNA internalization, plateau, and decrease of internal DNA content (Fig. 2 and 5). Denote, lag-period, introduction of foreign DNA in spermatozoa, saturation phase, and stage of DNA degradation were described in real experiments with washed sperm cells from different species [24]. I obtained the fast MHCII/DNA interaction by simulation, because the high value of k1 = 1 was used, which corresponded to a strong chemical binding. The same kinetic was found for human sperm cells [25]. In addition, the fine dynamics of DNA distribution between the participants–MHCII and CD4 proteins–in the course of sperm/DNA interaction was shown (Fig. 2 and 6ac). This hidden mechanism was not revealed in biochemical investigations. Related data were achieved when sperm cells from MHCII or CD4 knockout mice were used [17]. Thus, the simulation had a cognitive power.
Modeling with DNase activities demonstrated their significance in SMGT. The simulation showed that DNases in seminal fluid did not fully prevent the penetration of foreign DNA into spermatozoa (Fig. 3b). According to the model, the IF-1 protection leads to a significant decline of foreign DNA in sperm cells (Fig. 6c), which correlates with the particular inhibitor effect of IF-1 on CD4/DNA binding [14]. I achieved a total suppression of sperm/DNA interaction in the case of both IF-1 presence and DNase activity in the seminal fluid (Fig. 7a). In addition, the internal DNase activities evoked degradation of captured DNA (Fig. 4). Similar results were obtained in experiments with rabbit and bovine sperm, where DNase activity in cells was regulated by temperature [25]. The role of DNase activity in an apoptosis of sperm cells and in SMGT has been discussed [6, 26].
Artificial elimination of components from SP/DNA transport pathway in the virtual knockout experiments (Fig. 6) demonstrated the sufficient role of MHCII molecules in sperm/DNA interaction, as well as the participation of CD4 receptors in the internalization of DNA into spermatozoa. Surprisingly, the disconnection of MHCII and CD4 proteins (k2 = 0) had a more significant effect on DNA internalization than simple MHCII block (k1 = 0); the amounts of DNA in sperm cells at steady states were 0.08 and 0.33 c respectively (compare Fig. 6 b and a, red line). Broad variation in the content of MHCII on the mouse sperm surface was described by some authors and its role in SMGT was emphasized by Wu and coworkers [16]. The particular mechanism of DNA incorporation into mouse sperm cells with participation of MHCII and CD4 proteins was investigated by Spadafora’s team [17]. The mathematical model (1) described both phenomena.
In this work, I have used the system of first order linear ordinary differential equations–ODE model (1) that demonstrated the realistic behavior. Natural phenomena were described by variations on parameters in the model. However, the last experimental findings showed a complex network mechanism in foreign DNA processing by sperm cells [7, 10], some steps of which could be highly nonlinear and could lead to a more sophisticated behavior. I supposed the equilibrium state, which corresponds to sperm/DNA block, is a special case that might easily be disturbed and have unusual consequences. A large set of variations for parameters in the SMGT1 model caused ‘leaky’ states, where a part of exogenous DNA could penetrate sperm cells (Fig. 7b). These events could have taken place in nature and might have led to the foreign DNA penetration into spermatozoa. The percolation of reporter DNAs into ‘competent’ sperm cells was shown on the mussel Mitilus galloprovincialis [27, 28]. This kind of lateral gene transfer could be one of mechanisms for a compositional evolution of Eukaryotes [29].
Conclusion
Small variations in a number of MHCII and CD4 receptors on the sperm surface, as well as a change in IF-1 protecting protein and DNase activity in sperm could be a cause of sperm mediated foreign gene transfer. Combination of these factors can lead to important consequences, i.e. the extraneous DNA accumulation in internal parts of spermatozoa. The results of the simulation supported the hypothesis. This instability can be a source of sufficient genetic transitions in evolution.
Acknowledgements
The author is grateful to Irina Shchit and Anastasia Grigorenko for helpful discussions. This work was performed at the Physical and Biological Departments of the University of Freiburg, Germany.
References
1. Brackett BG, Baranska W, Sawicki W, Koprowski H. Uptake of heterologous genome by mammalian spermatozoa and its transfer to ova through fertilization. Proc Natl Acad Sci U S A. 1971. 68(2):353-7.
2. Arezzo F. Sea urchin sperm as a vector of foreign genetic information. Cell Biol. Intern. Rept. 1989. 13(4):391-404.
3. Lavitrano M, Camaioni A, Fazio VM, Dolci S, Farace MG, Spadafora C. Sperm cells as vectors for introducing foreign DNA into eggs: genetic transformation of mice. Cell. 1989. 57(5):717-23.
4. Lavitrano M, French D, Zani M, Frati L, Spadafora C. The interaction between exogenous DNA and sperm cells. Mol Reprod Dev. 1992. 31(3):161-9.
5. Giordano R, Magnano AR, Zaccagnini G, Pittoggi C, Moscufo N, Lorenzini R, Spadafora C. Reverse transcriptase activity in mature spermatozoa of mouse. J Cell Biol. 2000. 148(6):1107-13.
6. Sciamanna I, Barberi L, Martire A, Pittoggi C, Beraldi R, Giordano R, Magnano AR, Hogdson C, Spadafora C. Sperm endogenous reverse transcriptase as mediator of new genetic information. Biochem Biophys Res Commun. 2003. 312(4):1039-46.
7. Zoraqi G, Spadafora C. Integration of foreign DNA sequences into mouse sperm genome. DNA Cell Biol. 1997. 16(3):291-300.
8. Maione B, Pittoggi C, Achene L, Lorenzini R, Spadafora C. Activation of endogenous nucleases in mature sperm cells upon interaction with exogenous DNA. DNA Cell Biol. 1997. 16(9):1087-97.
9. Smith KR. The role of sperm-mediated gene transfer in genome mutation and evolution. Med Hypotheses. 2002. 59(4):433-7. Review.
10. Smith K, Spadafora C. Sperm-mediated gene transfer: applications and implications. Bioessays. 2005. 27(5):551-62. Review.
11. Prigogine I, Stengers I. Order out of Chaos - Man's new dialogue with nature. Bantam Books. Toronto. 1984. - 349 pages.
12. Kuznetsov AV, Kuznetsova IV. Mobile vector. 1998. - 189 pages. Retrieved date, from http://obomon.euro.ru/ Russian.
13. Kuznetsov AV. Toy SMGT // Alife Mutants Hackingsession on Systems and Organisms (AMHSO), Rule 110 Winter Workshop, Bielefeld, Germany, 6-13 March 2004. Retrieved date, from http://www.rule110.org/amhso/results/sperm-toy.pdf
14. Zani M, Lavitrano M, French D, Lulli V, Maione B, Sperandio S, Spadafora C. The mechanism of binding of exogenous DNA to sperm cells: factors controlling the DNA uptake. Exp Cell Res. 1995. 217(1):57-64.
15. Kuznetsova IV, Schit IYu, Kuznetsov AV. Deoxyribonuclease activity in the sperm of various animals in connection with peculiarities of fertilization. Russian Journal of Agricultural Biology. 1999. 2:77-9. Russian.
16. Wu GM, Nose K, Mori E, Mori T. Binding of foreign DNA to mouse sperm mediated by its MHC class II structure. Am J Reprod Immunol. 1990. 24(4):120-6.
17. Lavitrano M, Maione B, Forte E, Francolini M, Sperandio S, Testi R, Spadafora C. The interaction of sperm cells with exogenous DNA: a role of CD4 and major histocompatibility complex class II molecules. Exp Cell Res. 1997. 233(1):56-62.
18. Francolini M, Lavitrano M, Lamia CL, French D, Frati L, Cotelli F, Spadafora C. Evidence for nuclear internalization of exogenous DNA into mammalian sperm cells. Mol Reprod Dev. 1993. 34(2):133-9.
19. Kuznetsov AV, Kuznetsova IV, Schit IY. DNA interaction with rabbit sperm cells and its transfer into ova in vitro and in vivo. Mol Reprod Dev. 2000. 56(2 Suppl):292-7.
20. Maione B, Lavitrano M, Spadafora C, Kiessling AA. Sperm-mediated gene transfer in mice. Mol Reprod Dev. 1998. 50(4):406-9.
21. Kuznetsov AV, Kaurova SA, Kuznetsova IV. Transfection of endometrium after rabbit insemination by transformed sperm cells. Model of DNA internalization into spermatozoon. Russian Journal of Human Reproduction. 1998. 4(6):29-33. Russian.
22. Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in FORTRAN 77: The Art of Scientific Computing Vol 1 (2nd ed). Cambridge University Press. 1996. - 1486 pages.
23. Kuznetsov AV, Sigaeva VA, Kuznetsova IV, Schit IYu. Rabbit spermatozoa are capable binding of foreign DNA. Russian Journal of Human Reproduction. 1996. 1:7-10. Russian.
24. Castro FO, Hernandez O, Uliver C, Solano R, Milanes C, Aquilar A, Perez A, de Armas R, Herrera L, de la Fuente J. Introduction of foreign DNA into the spermatozoa of farm animals. Theriogenology. 1990. 34(6):1099-1110.
25. Sigaeva VA, Kuznetsova IV, Kuznetsov AV. Exogenous DNA binding by spermatozoa. Russian Journal of Human Reproduction. 1996. 2:18-20. Russian.
26. Spadafora C. Sperm cells and foreign DNA: a controversial relation. Bioessays. 1998. 20(11):955-64. Review.
27. Kuznetsov AV, Pirkova AV, Dvorianchikov GA, Panfertsev EA, Gavriushkin AV, Kuznetsova IV, Erokhin VE. Study of the transfer of foreign genes into mussel Mytilus galloprovincialis Lam. eggs by spermatozoa. Ontogenez. 2001. 32(4):309-18. Russian.
28. Guerra R, Carballada R, Esponda P. Transfection of spermatozoa in bivalve molluscs using naked DNA. Cell Biol Int. 2005. 29(2):159-64.
29. Watson RA. Compositional Evolution: The Impact of Sex, Symbiosis, and Modularity on the Gradualist Framework of Evolution. (Vienna Series in Theoretical Biology) A Bradford Book. 2006. 324 p.
Appendix
The program SMGT1 in Matlab7.