Ryan N. Willhite Week 9
From OpenWetWare
Jump to navigationJump to search
Week 9 Work Day
- Notes/brainstorming
- Bedrock
- Retrieving sequences from those compared in HIV Evolution Project.
- Amino acid sequences instead of nucleic
- Viewing subjects 5,6,7,10, and 13.
- Utilizing Protein tools on workbench in order to upload these sequences.
- After uploading, doing a multiple sequence alignment to compare
- Comparisons include: (6-13, 6-5, 13-5, 7-10, 7-5, 10-5)
- If time and interesting data, comparisions will be made between all subjects to cover all areas.
- If enough time, time points will also be looked at in the most interesting of all these sequences in order to determine the changes in structure atthree different time points.
- Questions
- ORFing allows one to go from DNA sequences to proteins sequences.
- www.ncbi.nlm.nih.gov/gorf/gorf.html
- Possible experiments and tools
- ProtParam
- Predict the physico-chemical properties of a protein
- Scan Prosite
- analyzing post translational modifications
- retrieving a 3D structure
- Overall, since the DNA sequences failed to give any significant evidence perhaps protein confirmations and information can help provide more information on the categorization of progression.
Workbench Information
Mutliple Sequence Alignment Results
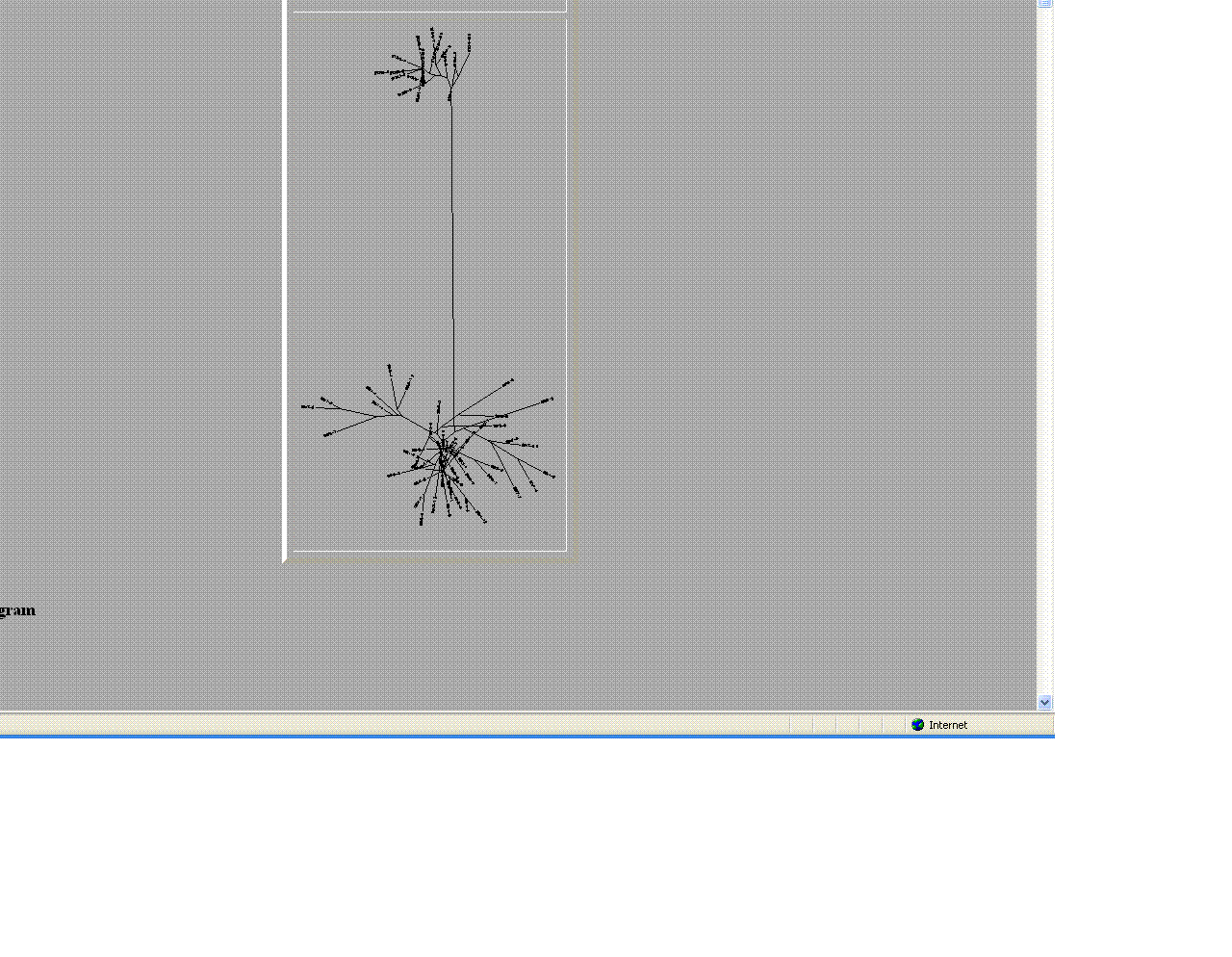
- Comparison of Subject 6 and 13
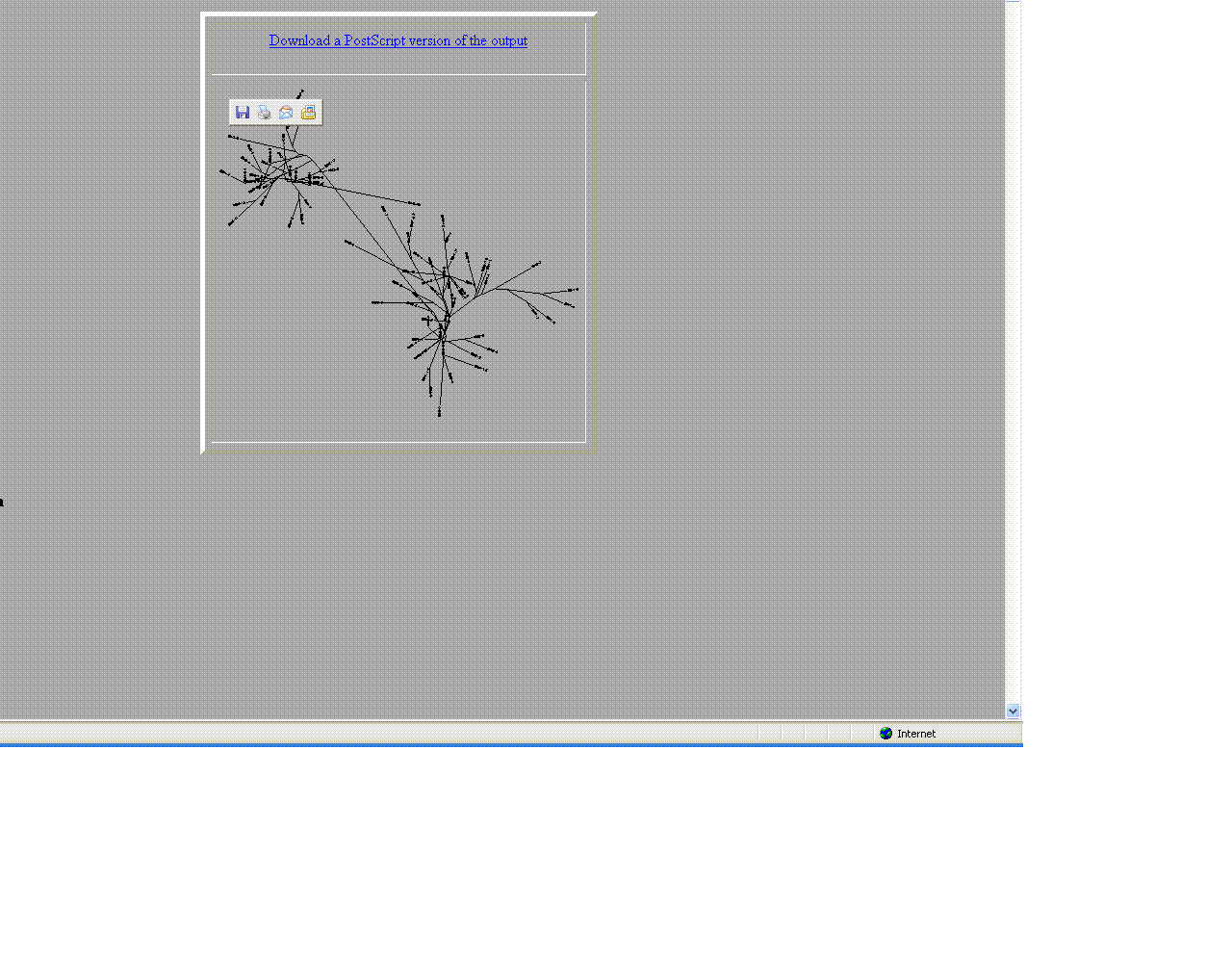
- Comparison of Subject 6 and 5
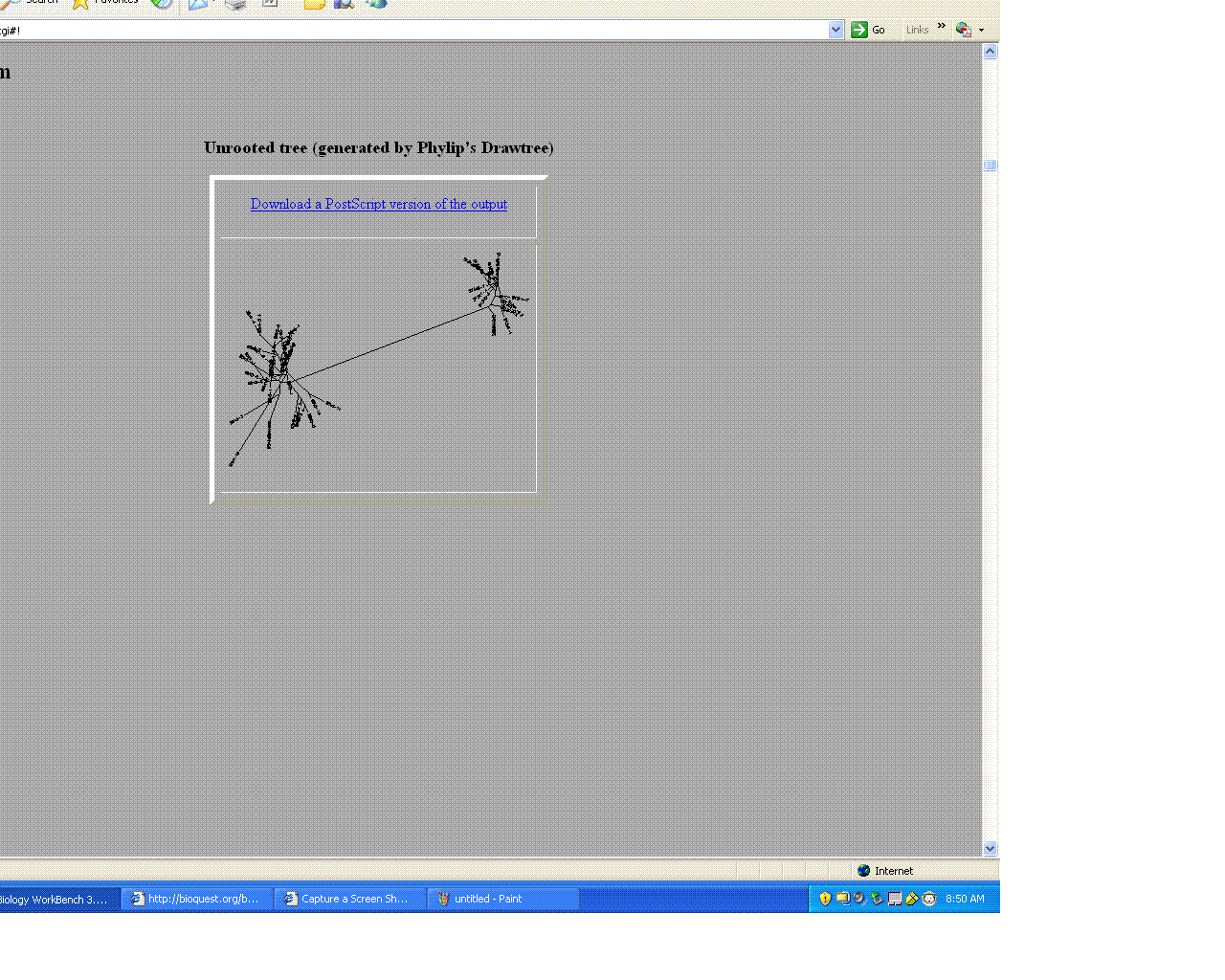
- Comparison of Subject 13 and 5
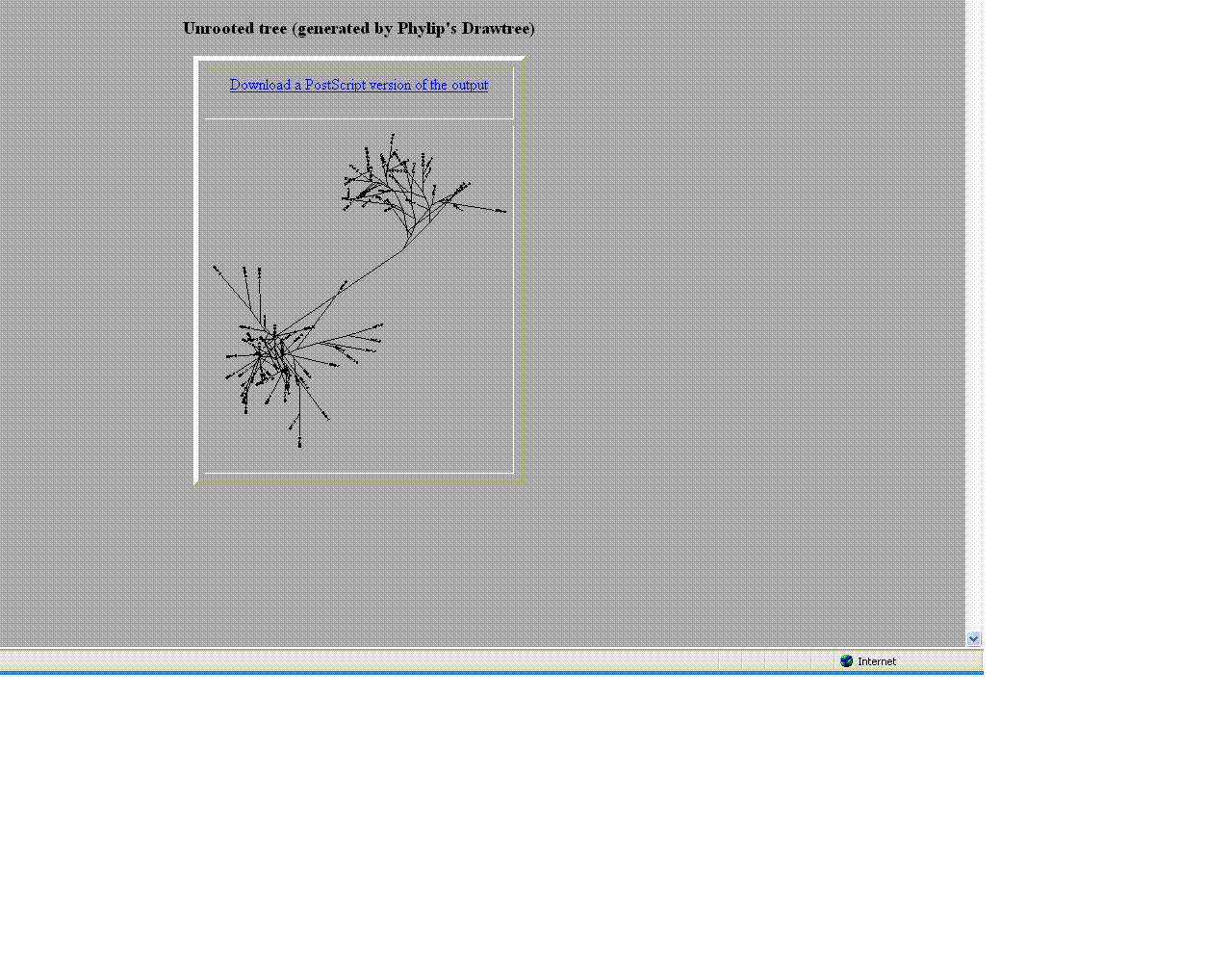
- Comparison of Subject 6 and 7
Comments/Notes
- The trees helped give a visual as the to similarities and differences between these moderate and non- progressors.
- As seen above the non- progressors were quite different than those of the moderate progressors.
- Two moderates being compared were very similar in sequences
- This may suggest that looking at protein sequences are a more efficient way of categorizing.
- This leads to a curiosity in structure
- Structures between moderate subjects such as subject 7. subject 6, and subject 5
- With results suggesting 7 maybe more like a rapid progressor than moderate progressors, these structures will b viewed as well
- Subject 7/10 will be compared, with hopefully similarities that can potentially put 7 in the rapid progressor category according to a similarity in structure.
ProtParam
- Compare Subjects 5/6/7/10
- www.expasy.org/tools/#primary
- ProtParam tools
- Entered amino acid sequences
Results
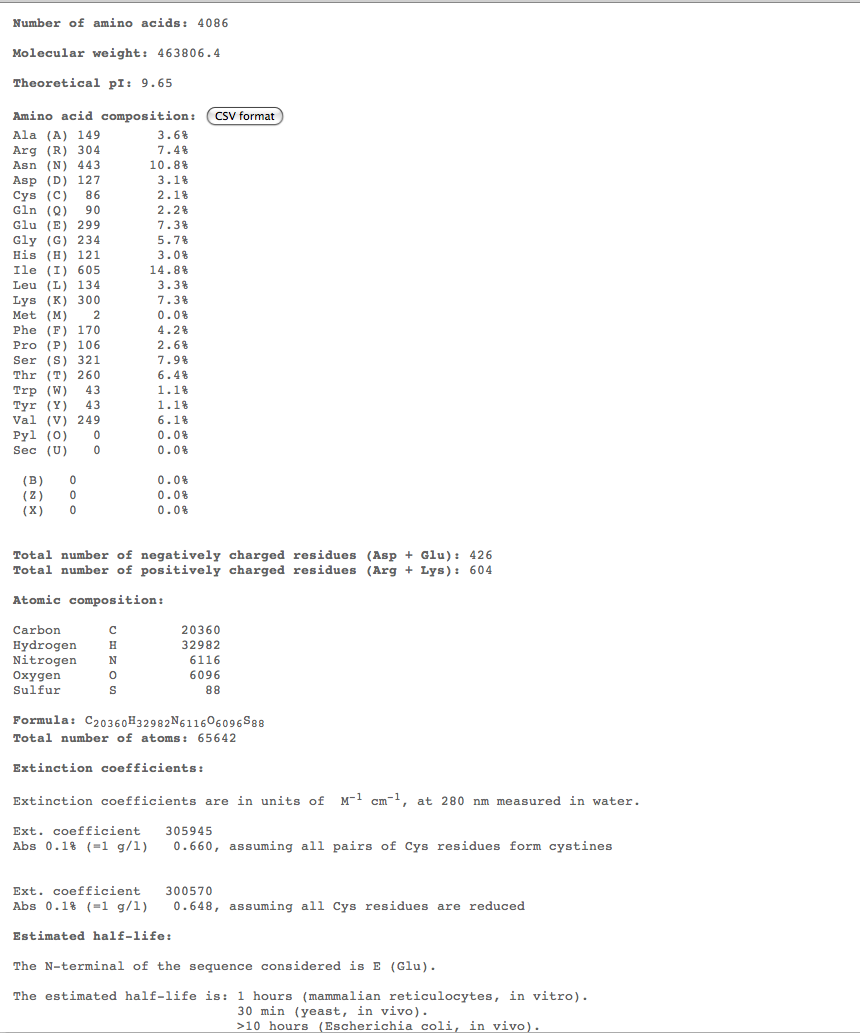
- Subject 5
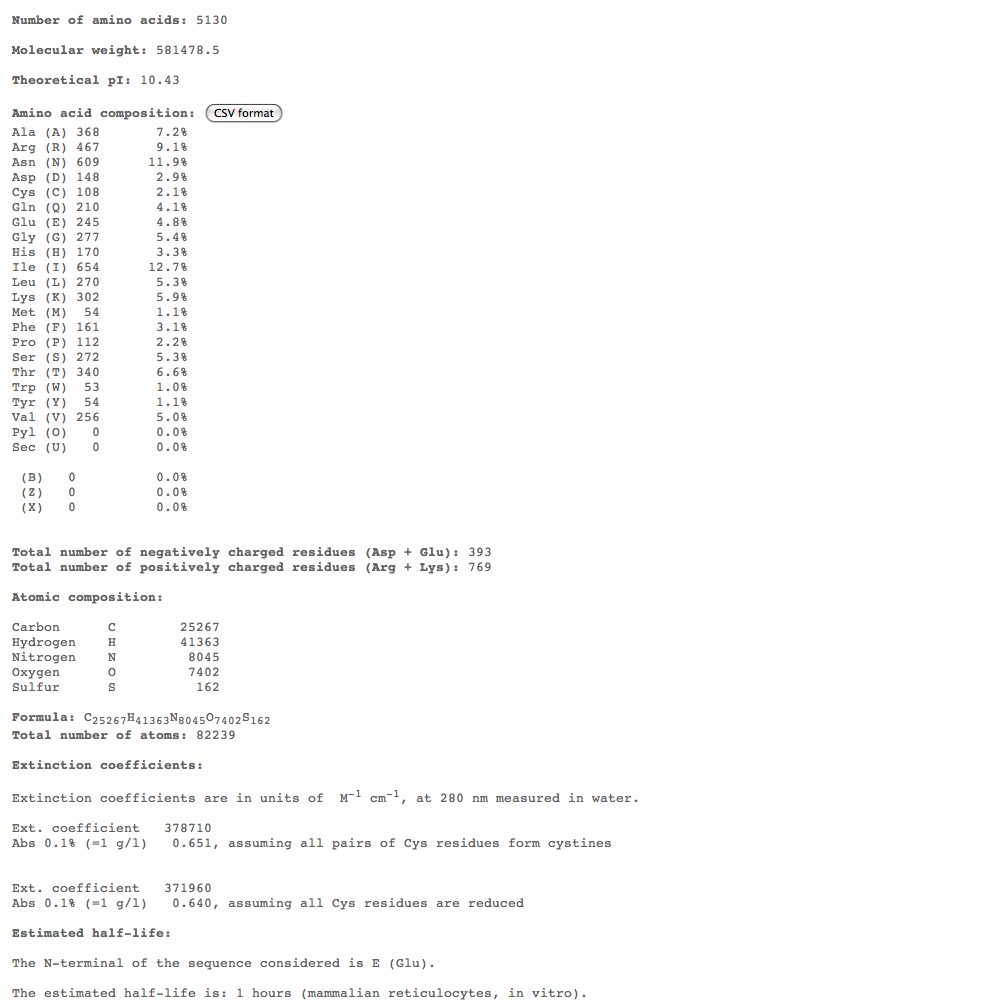
- Subject 6
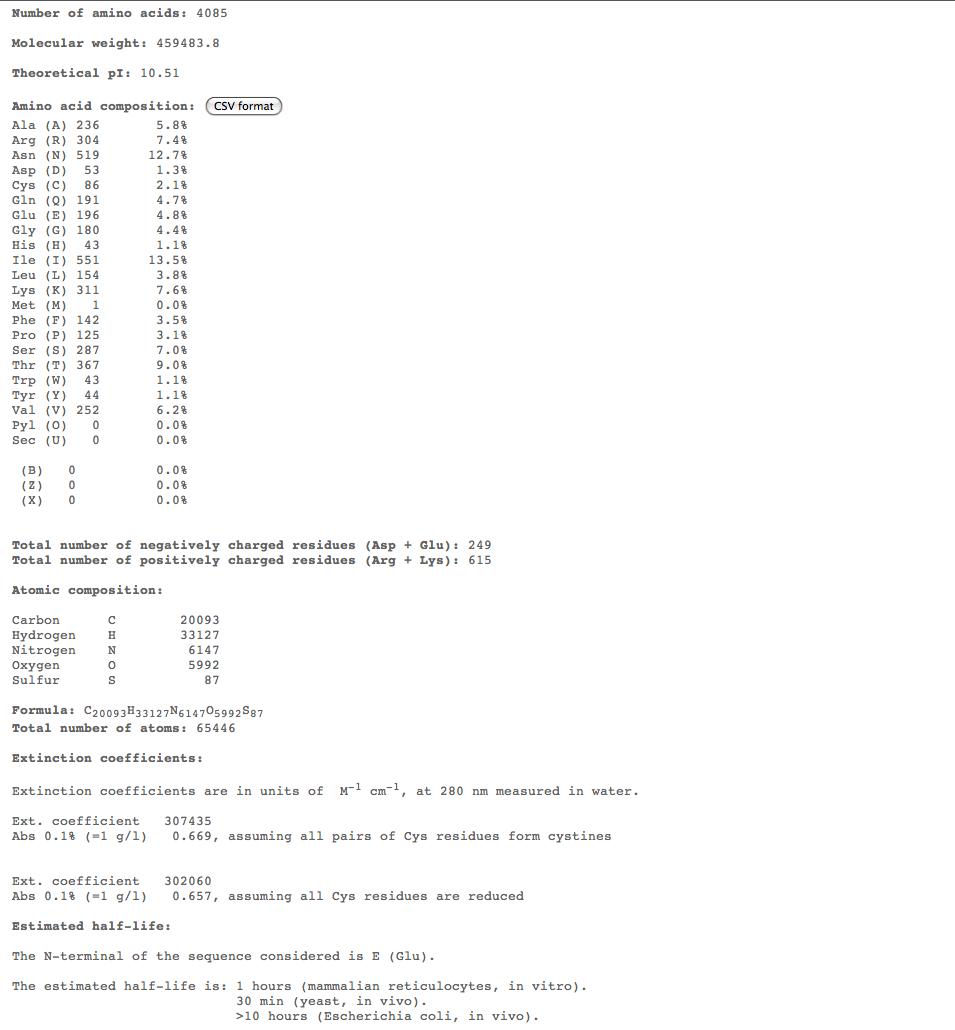
- Subject 7
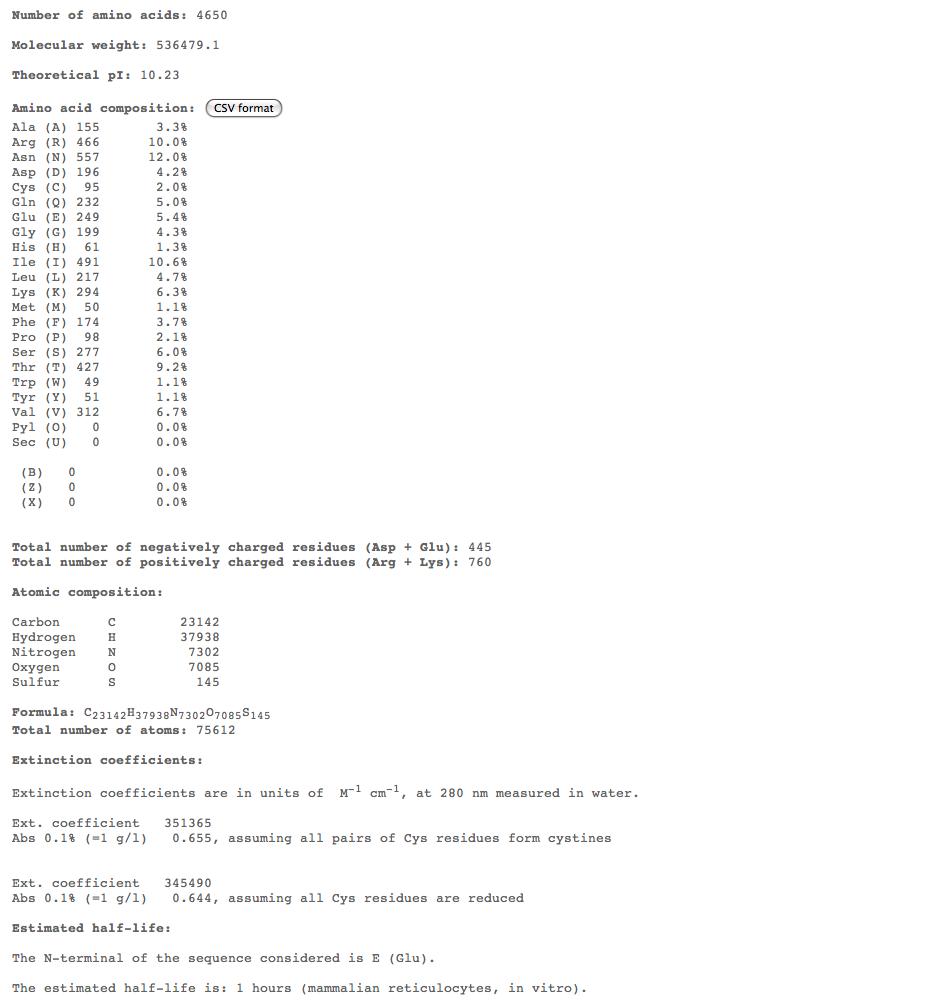
- Subject 10
Comments
- The highest percent of amino acids found in the moderates 5,6, and 7 is (Ile)
- THe highest percent of amino acids found in the rapid 10 to compare is (Asn)
- Looking at these compositions along with protein structure seems to be a more efficient way of categorizing progression. The difference in amino acid composition suggests a role in type of progression.
Paper/ppt
<biblio>
- Paper1 pmid=18215327