Robert W Arnold Week 8
From OpenWetWare
Jump to navigationJump to search
Electronic Lab Notebook 8
In-Class Protein Activity
Chapter 2
- Started off with Bioinformatics for Dummies. Logged into UniProt site.
- Determined AA sequence of gp120, here is the FASTA format: >sp|P04578|33-511
KLWVTVYYGVPVWKEATTTLFCASDAKAYDTEVHNVWATHACVPTDPNPQEVVLVNVTEN FNMWKNDMVEQMHEDIISLWDQSLKPCVKLTPLCVSLKCTDLKNDTNTNSSSGRMIMEKG EIKNCSFNISTSIRGKVQKEYAFFYKLDIIPIDNDTTSYKLTSCNTSVITQACPKVSFEP IPIHYCAPAGFAILKCNNKTFNGTGPCTNVSTVQCTHGIRPVVSTQLLLNGSLAEEEVVI RSVNFTDNAKTIIVQLNTSVEINCTRPNNNTRKRIRIQRGPGRAFVTIGKIGNMRQAHCN ISRAKWNNTLKQIASKLREQFGNNKTIIFKQSSGGDPEIVTHSFNCGGEFFYCNSTQLFN STWFNSTWSTEGSNNTEGSDTITLPCRIKQIINMWQKVGKAMYAPPISGQIRCSSNITGL LLTRDGGNSNNESEIFRPGGGDMRDNWRSELYKYKVVKIEPLGVAPTKAKRRVVQREKR
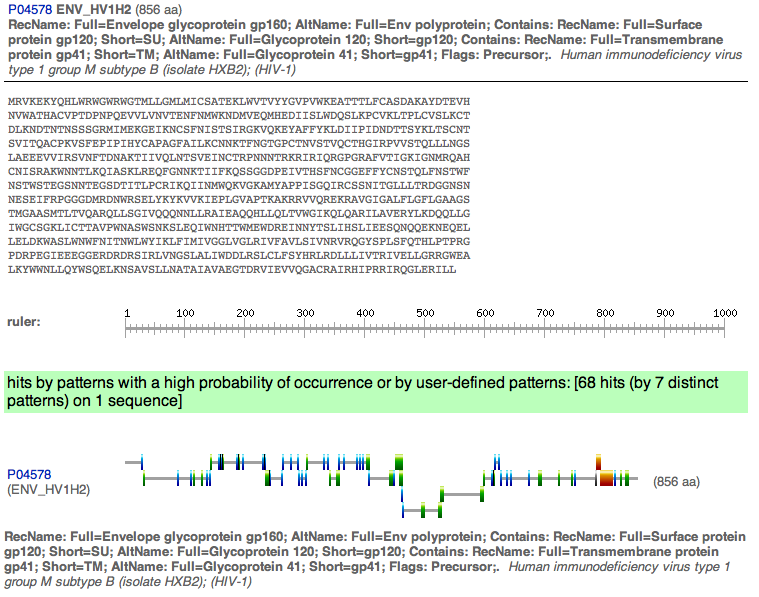
- Here is a picture of the gp120 sequence.
- Saved list and FASTA format of gp120 |P04578|.
Chapter 4
- Here is a link to the SWISS-PROT entry.
- Here is the entry information:
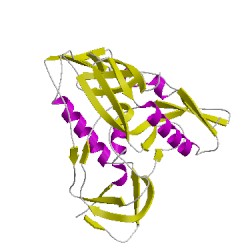
- Here is a picture of gp120:
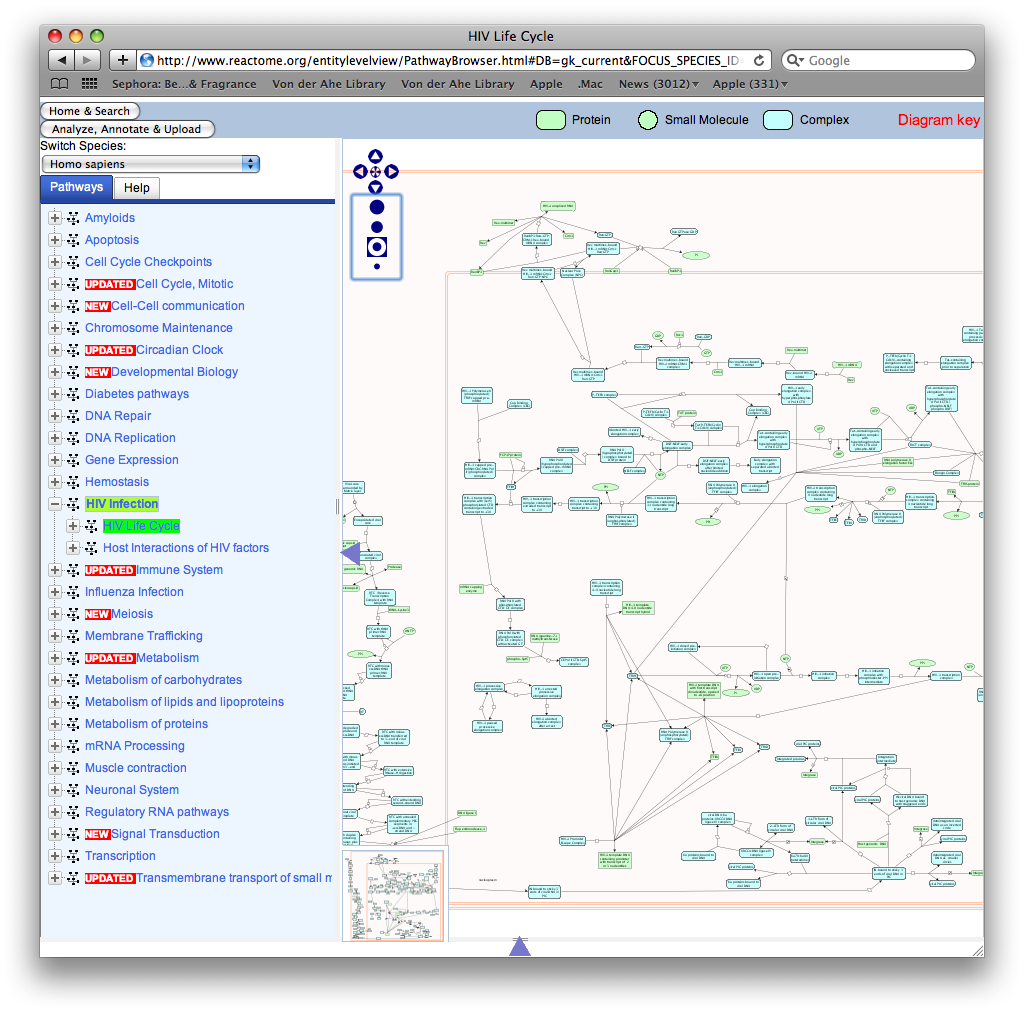
- Found a great place for enzyme and pathway information in Reactome. Here is an image of the information. As you can see on the left you can go through gp120 function and HIV function, etc.
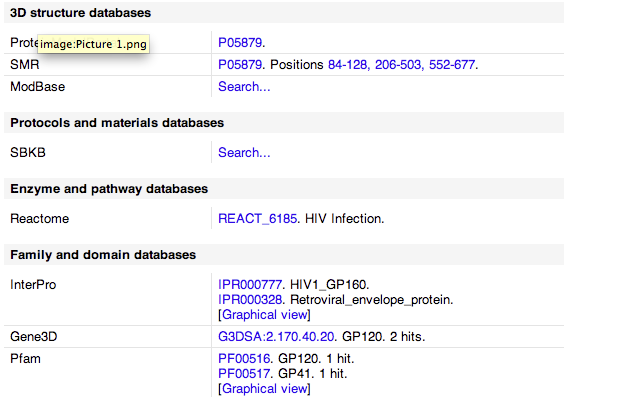
- Here is a Gene3D image of gp120.
- After this I determined the references at the bottom of the page.
- Looked at cross references which had links to other sites with protein information which included some of the following.
Chapter 5
- Opened up the NCBI link and entered in FASTA format of gp120 to use the ORF Finder. Here are the results:
- The DNA sequence is seen the the 6 bars.
- ORF Finder allows one to find coding regions with DNA.
Chapter 6
- Opened up ExPASy website.
- Selected ProtParam tool at top of site.
- Entered my sequence into tool
- Looked at list of different selectable regions, chose third one down from top. gp120 Surface protein 33-511.
- Here is screen shot of ProtParam output.
- ProtParam info showed many important qualities of the protein like molecular weight, aa composition, atomic composition, half-life, etc.
- The next section involved looking for transmembrane segments using Protscale and TMHMM.
- The sequence from 512-856 of the transmembrane protein gp 41 was selected.
- Here are the results showing:
- Then used the link provided for the TMHMM tool, www.cbs.dtu.dk/services/TMHMM-2.0. Imported FASTA sequence.
- Here is results of TMHMM, spiked areas show where transmembrane region most likely found.
- Then the ScanProsite was used.
- Here are the results, with the do not scan profiles box checked and exclude motifs unchecked.
- Next used was InterProScan to find the domains and here are the results.
HIV Structure Research Project
- Zeb, Alex, and I will be working together again.
- Planning on developing a question and researching the area of V3 conformation difference between subjects with and without AIDS, look to see if conformational change took place after AIDS set in.
- Will look at individual subjects to look for conformation changes, hope to see similarities between closely related subjects, related by close dN/dS numbers, CD 4 T cell counts etc.
- Expecting to see conformational change up until AIDS sets in. Expect this to be the best conformation for that certain virus.
- Going to look at all of the rapid progressors with all their sequencing and then one or two others for more comparison. Rapid progressors will be used because we have all of their sequences and we will choose one other moderate progressor and non progressor for comparison.
- The subjects we are going to pick are 1, 3, 4, 10, 11, 15, and then moderate progessor 6 and 13.
Links
- Robert W Arnold Week 2
- Robert W Arnold Week 3
- Robert W Arnold Week 4
- Robert W Arnold Week 5
- Robert W Arnold Week 6
- Robert W Arnold Week 7
- Robert W Arnold Week 8
- Robert W Arnold Week 9
- Robert W Arnold Week 10
- Robert W Arnold Week 11
- Robert W Arnold Week 12
- Robert W Arnold Week 14
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 14 Assignment
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 10
- Class Journal Week 11
- Class Journal Week 12
- Class Journal Week 14