Biomod/2012/HKBU/BU Magician:Methods
<html> <style> .container{background-color: #FFFFFF; margin-top:0px}
- column-content {width: 0px; float: left; margin: 0 0 0 0;padding: 0;}
.firstHeading {display:none; width:0px;}
- column-one {display:none; width:0px;background-color: #FFFFFF;}
- globalWrapper{width: 920px; background-color: #FFFFFF; margin-left: auto;margin-right: auto;
-moz-box-shadow: 0px 0px 15px #ccc; -webkit-box-shadow: 0px 0px 15px #ccc; box-shadow: 0px 0px 15px #ccc;}
- content{ margin: 0 0 0 0; align: center; padding: 12px 12px 12px 12px; width: 800px;background-color: #FFFFFF; border: 0;}
- bodyContent{ width: 800px; align: center; background-color: #FFFFFF;}
- column-content{width: 800px;background-color: #FFFFFF;}
</style> </html> <html xmlns="http://www.w3.org/1999/xhtml" xml:lang="en" lang="en"> <head> <meta http-equiv="Content-Type" content="text/html; charset=UTF-8" /> <title>jOuery</title> <link rel="stylesheet" href="style.css" /> <style> .menu { height: 45px; display: block; } .menu ul { list-style: none; padding: 0; margin: 0; } .menu ul li { float: left; overflow: hidden; position: relative; text-align: center; line-height: 45px; } .menu ul li a { position: relative; display: block; width: 128px; height: 45px; font-family: Arial; font-size: 11px; font-weight: bold; letter-spacing: 1px; text-transform: uppercase; text-decoration: none; cursor: pointer; } .menu ul li a span { position: absolute; left: 0; width: 128px; } .menu ul li a span.out { top: 0px; } .menu ul li a span.over, .menu ul li a span.bg { top: -45px; }
- menu2 { background:#45A8DF; }
- menu2 ul li a { color:#FFF; }
- menu2 ul li a span.over { background: #A6DD00; color:#333; }
- menu2 ul li.nav1 a span.over { background: #fea274; }
- menu2 ul li.nav2 a span.over { background: #b0bbba; }
- menu2 ul li.nav3 a span.over { background: #a3f091; }
- menu2 ul li.nav4 a span.over { background: #86dbf9; }
- menu2 ul li.nav5 a span.over { background: #e0caf0; }
- menu2 ul li.nav6 a span.over { background: #9dace9; }
- menu2 ul li.nav7 a span.over { background: #FFE66F; }
</style> <script type="text/javascript" src="http://ajax.googleapis.com/ajax/libs/jquery/1.3/jquery.min.js"></script> <script language="javascript"> $(document).ready(function() {
$("#menu2 li a").wrapInner( '' );
$("#menu2 li a").each(function() { $('' + $(this).text() + '' ).appendTo( this ); });
$("#menu2 li a").hover(function() { $(".out",this).stop().animate({'top':'45px'},200); $(".over",this).stop().animate({'top':'0px'},200);
}, function() { $(".out",this).stop().animate({'top':'0px'},200); $(".over",this).stop().animate({'top':'-45px'},200); });
});
</script> </head> <body>
</body> </html> <html>
Methods and Techniques
</html>
Techniques for Experiment
Cell culture
Cell culture refers to the complex process by which cells are grown under controlled conditions, generally outside of their natural environment. The culturing of cells derived from multi-cellular eukaryotes, especially animal cells are now most commonly used.
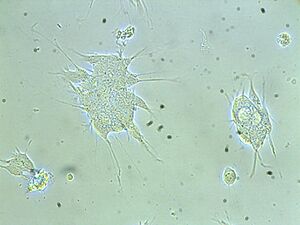
For a cell culture, cells need to be isolated from tissues for ex vivo culture. To maintain cells in culture, the appropriate temperature and gas mixture (typically, 37°C, 5% CO2 for mammalian cells) in a cell incubator, the appropriate growth medium (vary in pH, glucose concentration, growth factors, and the presence of other nutrients) and the plating density (number of cells per volume of culture medium) are all very crucial in culture system. In our experiment, SH-SY5Y cells from human brain are isolated for cell culture. The cell incubator condition is as usual, 37°C and 5% CO2 and the growth medium is DMEM / F12 Media [Dulbecco's Modified Eagle Medium (DMEM) : Nutrient Mixture F-12 (Ham's) = (1:1)]. In our experiment, cells grow in adherent cultures, requiring a surface such as tissue culture plastic.
As cells generally continue to divide in culture, they generally grow to fill the available area or volume. This can generate several issues:
1.Nutrient depletion in the growth media
2.Accumulation of apoptotic/necrotic (dead) cells
3.Cell-to-cell contact can stimulate cell cycle arrest, causing cells to stop dividing, known as contact inhibition.
4.Cell-to-cell contact can stimulate cellular differentiation.
To solve the problem above, several manipulations could be done: media changes, passaging cells, transfection and transduction.
Media changing refers to the media being removed directly by aspiration, and then is replaced.
Passaging (also known as subculture) involves transferring a small number of cells into a new vessel. Cells can be cultured for a longer time if they are split regularly, as it avoids the senescence associated with prolonged high cell density. For adherent cultures, cells first need to be detached, commonly done with a mixture called trypsin-EDTA. In our experiment, passaging is done every week and the common number used to transfer to a new vessel is 1x106 cells /flask.
Manipulating cells involves the introduction of foreign DNA by transfection, which is often performed to cause cells to express a protein of interest. More recently, the transfection of RNAi constructs have been realized as a convenient mechanism for suppressing the expression of a particular gene/protein. In our experiment, siRNA is transfected to the cells to suppress the expression of the interested protein, NR2B protein.
Immunofluorescence
Immunofluorescence uses the specificity of antibodies to their antigen in order to target fluorescent dyes to specific target biomolecule within a cell, and therefore allows visualization of the distribution of the target molecule in the sample. In this experiment, indirect immunofluorescence method is used. The unlabeled first (primary) antibody specifically binds the target molecule, and the secondary antibody, which carries the fluorophore, recognizes the primary antibody and binds to it. This provides signal amplification by increasing the number of fluorophore molecules per antigen.
In our experiment, the primary antibody is rabbit NR2B antibody with 2% normal goat serum, and the secondary antibody is NR2B Fluorescein(Alexa 488)-conjugated goat anti-rabbit secondary antibody.
Confocal microscopy
Confocal microscopy, as an optical imaging technique, uses point illumination and a spatial pinhole to eliminate out-of-focus light in specimens, so that it could be used to increase optical resolution and contrast of a micrograph. Comparing with the traditional wide-field fluorescence microscope which will excite all parts of the specimen n the optical path, a confocal microscope uses point illumination and a pinhole in an optically conjugate plane in front of the detector to eliminate out-of-focus signal. In this way, the image’s optical resolution could be increased because only light produced by fluorescence very close to the focal plane can be detected.
In this experiment, confocal microscopy is used to take photos of cover slips which have been given cell treatments and the primary and secondary antibodies have been added. Because the secondary antibodies are with fluorescence, we hope to analyze the intensity of fluorescence to see the proteins the cell expressed.
Methods
Conjugations

Goals:
To synthesize siRNA with nanoparticle solutions and wait until conjugation for next part of treatment
Methods:
Nanoparticles solutions: Aqueous dispersions of magnetite nanoparticles were prepared according to Massart’s method [R. Massart, IEEE Trans. Magn. 1981, MAG-17, 1247.], based on the co-precipitation of ferrous and ferric ion solutions (1:2 molar ratio).
Conjugations
Dilute iron nanoparticle solution to the concentration wanted. Then add the add siRNA to nanoparticle solution. Store conjugated solutions in the refrigerator for 24 hours for further treatment.
*Calculation should have been done according to the final siRNA and nanoparticle concentration wanted
Treatment
Goals:
To give cells treatments with conjugated siRNA and nanoparticle solutions
Methods:
Cell used: SH-SY5Y neuroblastoma cells
Cell seeding: Before seed the cells into the well, autoclaved cover slips are put into each well of a 4-well plate and 500μl Poly-D-lysine is for improving adherence of the cells to the cover slips. After cells are cultured into wells, it will be put into the incubator overnight.
Cell treatment:
DMEM/F-12 complete medium is removed and replaced by DMEM/F-12 serum free medium Perform the treatment in different wells and then they will be incubated for 24 hours.
For the design of the experiment, several groups should be concluded:
a). iron-siRNA group (For each concentration, perform 2 sets, one with magnetic field and one without magnetic field)
b). iron group
c). siRNA group
d). control group (only medium)
<html>
<img src="http://openwetware.org/images/f/f8/HKBUTABLE2.jpg" height="429" width="800"/>
</html>
Antibodies Adding and Slide Preparation
Goals: To add primary antibodies and secondary antibodies(with fluorescence) to the treated cells and prepare slides for confocal
Methods:
In our experiment, the primary antibody is rabbit NR2B antibody with 2% normal goat serum, and the secondary antibody is NR2B Fluorescein (Alexa 488)-conjugated goat anti-rabbit secondary antibody.
Primary antibody containing solution(concentration: 1:1000):
a). NR2B antibody
b). PBS with 0.1% Triton X
c). 2% normal goat serum (NGS)
Secondary antibody containing solution(concentration: 1:500):
a). Fluorescein-conjugated goat anti-rabbit secondary antibody
b). PBS
Fluoresence Detection and Data Analysis
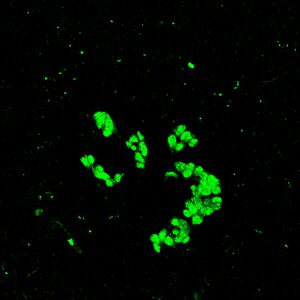
Goals: To take photos of immunostained cells under confocal microscope and further analyze the effect of nanoparticle with SPSS one-way function
Methods:
1. Observed the slides under Laser scan Confocal Microscope (FluoViewTM FV1000, Olympus; excitation of Alexa 488) and capture 3 images of each slides.
2. Measure the signal intensity for cells on each image using software (Metamorph).
3. Collect data of the each set of treatment and use SPSS to calculate changes of intensity and the significance of the changes.
For data analysis:
Significance:
We use SPSS software, one-way anova to test the significance of the changes. Statistical significance is a statistical assessment of whether observations reflect a pattern rather than just chance. The fundamental challenge is that any partial picture of a given hypothesis, poll or question is subject to random error. Popular levels of significance are 10% (0.1), 5% (0.05), 1% (0.01), 0.5% (0.005), and 0.1% (0.001). If a test of significance gives a p-value lower than the significance level α, the null hypothesis is rejected. Such results are informally referred to as 'statistically significant'. Graphically, statistical significance is indicated by the use of star symbols (*). The number of stars usually indicates the significance level: one star (*) for 0.05, two (**) for 0.01, and three (***) for 0.001 or 0.005.
Decrease Percentage Calculation:
Here, we use the decrease percentage to see the trend of treatment effect roughly.
Decrease Percentage =(Intensity of control group - Intensity of cell treatment) / Intensity of control gourp
Standard Protocol
1st Day Solution preparation& Seeding Cell
Solution preparation
Nanoparticle solution preparation
For nanoparticle stock solution: 20 mL of aqueous 1 M FeCl3 (97%, Aldrich) and 5 mL of 2 M FeSO4• 7H2O (99%, Riedel-de-Haen) in 2 M HCl were added to 250 mL of 0.7 M NH4OH (28–30%, Aldrich) under rapid mechanical stirring. Stirring was allowed to continue for 30 min, and then the black solid product was allowed to precipitate. The sediment was redispersed in 50 mL distilled water, and subsequently three aliquots of 10 mL tetramethylammonium hydroxide solution (10% in water, Aldrich) (1 M) were added, again with rapid stirring. Finally, water was added to the dispersion up to a total volume of 250 mL. The magnetite nanoparticles (which were partially oxidized to maghemite) were further washed as follows: 4 mL of magnetic-nanoparticles solution was diluted with 36 mL distilled water containing 50 lL HCl (0.01 M), centrifuged, and redispersed in pure water.
Silica-Coated Magnetite/Maghemite Particles (Fe3O4/c-Fe2O3@SiO2): To a mixture of 4.85 mL NH4OH (28 %), 28.8 mL H2O, 27.5 mL EtOH, and 6.22 mL of the previously washed magnetic nanoparticles in aqueous solution, an ethanolic solution of TEOS (98%, Aldrich) (2 mL of TEOS in 30 mL of EtOH) was added while mechanically stirring. After mixing, the final volume was 100 mL and the concentration of TEOS, NH4OH, and H2O were, respectively, 0.25, 0.35, and 21.4 M. The hydrolysis and condensation of TEOS onto the magnetic nanoparticles was completed in 4 h. The formed particles were centrifuged to eliminate excess reactants and redispersed in 50 mL of pure water. These core/shell magnetic silica spheres had an average diameter of 170 ± 10 nm.
250μl of nanoparticle stock solution (conc.=40mg/ml) is diluted with 1750μl HBSS (Concentration: 5mg/ml) Solution with concentration 5 times as working concentration is prepared here. <html> <img src="http://openwetware.org/images/5/57/Hkbu2012table1.jpg" width="750px" height = "170" /> </html>
SiRNA containing solution preparation
For each well, 300μl of nanoparticle solution is needed for treatment.
For SiRNA containing nanoparticle solution
60μl is added to 240 μl serum free medium to dilute 5 times.
Total volume: 60x 2(duplicates) x2(with magnetic field/without magnetic field)=240μl
Take in account the pipetting error, 240μl -> 320μl for each concentration solution containing nanoparticles
SiRNA stock concentration:40μM
SiRNA concentration needed for treatment 200nM
SiRNA concentration for nanoparticle solution preparation: 1000nM
<html><img src="http://openwetware.org/images/5/58/HKBU2012TABLE4.jpg" height="158" width="800"/></html>
( *Remarks:The final concentration of SiRNA containing nanoparticle solution is not calculated here.)
40*1000*X=1000(concentration)*320 X=8
Remarks: if under this condition, the nanoparticle is too concentrated for conjugation.
For SiRNA containing incomplete medium solution (experiment 9,10)
Volume of SiRNA containing medium= 60μlx2 (duplicates) x 2(experiment 9,10) =240μl
Take in account of pipetting error, 240μl-> 320μl <html><img src="http://openwetware.org/images/f/fe/HKBUTABLE6.jpg" height="107" width="664"/></html>
Conjugation
After adding SiRNA to both nanoparticle and HBSS solution, the tubesare stored in 4 centigrade refrigerator to allow conjugating overnight for 24hrs.
Solution preparation
Precoat the 4 well plate
Seeding cells After cell count, calculate the volume should be added into each well according to the cell number of 5x104 cells/well. Place in incubator overnight to allow cell growth
2nd Day Solution preparation& Seeding Cell
Cell Treatment
Solutions Adding (to wells)
Decant medium
Add corresponding solution to the wells <html> <img src="http://openwetware.org/images/f/f2/HKBUTABLE7.jpg" height="450" width="800"/> </html>
SiRNAdelivary=
After adding cells to solutions according to the table above, the wells are stored in incubator for 24hrs.
3rd Day Antibody Adding
Primary Antibody Adding
1.Discard all medium in the wells.
2.Washed the cells gently with PBS 3 times.
3.Fix the cells using 3% paraformaldehydefor 30 minutes at room temperature.
4.Washed the cells gentlywith PBS 3 times.
5.Add primary antibody containing solution(350μl/well).
6.Incubate the cells for 2 hours at room temperature or overnight under 4°C refrigerator.
Primary antibody containing solution(concentration: 1:1000):
a). NR2B antibody
b). PBS with 0.1% Triton X
c). 2% normal goat serum (NGS)
1ml primary antibody containing solution = 1μl NR2B antibody + 20ul NGS + 979ul 0.1% TritonX PBS
350μl primary antibody containing solution/well X 26 wells =9100μl primary antibody containing solution
Primary antibody containing solution preparation:
10ml primary antibody containing solution
= 10μl NR2B antibody + 200ul NGS + 9790ul 0.1% TritonX PBS
Secondary Antibody Adding
7. Discard all primary antibody containing solution.
8. Wash with PBS three times.
9. Add secondary antibody containing solution(350μl/well).
10. Incubate the cells for 2 hours at room temperature.
Secondary antibody:
Fluorescein(Alexa 488)-conjugated goat anti-rabbit secondary antibody
Secondary antibody containing solution(concentration: 1:500):
a). Fluorescein-conjugated goat anti-rabbit secondary antibody
b). PBS
1ml secondary antibody concentration = 2μl of goat anti rabbit Alexa 488 + 998μl PBS (bench use PBS)
350μl secondary antibody containing solution/well X 26 wells = 9100μl secondary antibody containing solution
Secondary antibody containing solution preparation:
10ml secondary antibody containing solution
=20μl of goat anti rabbit Alexa 488 + 9980μl PBS (bench use PBS)
Slide Preparation
11.Discard all secondary antibody containing solution and wash with PBS three times.
12.Add one drop mounting medium (DAKO) onto a clean slide.
13.Coverslips with immunostained cells were mounted on the clean slide (without bubbles).
4th Day Images Taking and Data Analysis
1. Observed the slides under Laser scan Confocal Microscope(FluoViewTM FV1000, Olympus; excitation of Alexa 488).
2. For each cover slip, take 3 photos under different parts of the cover the slip.
3. Collect the photos and measure the signal intensity for each cell using software (Metamorph).
4. Collect data from Metamorph and use SPSS to calculate changes of intensity and the significance of the changes.
<html> <a href="http://openwetware.org/index.php?title=Biomod/2012/HKBU/BU_Magician:Methods&action=edit">edit</a> </html>