BioMicroCenter:DNA LIB
| HOME -- | SEQUENCING -- | LIBRARY PREP -- | HIGH-THROUGHPUT -- | COMPUTING -- | DATA MANAGEMENT -- | OTHER TECHNOLOGY |
The BioMicro Center offers a broad variety of methodologies for preparing DNA libraries for sequencing. As will all of our standard prep methods, pricing includes quality control prior and following prep, along with at least one re-prep for samples that fail. For large batches of samples or 16S/amplicon, please visit our High-Throughput DNA library preparation page.
| Summary | Method | Input | Insert Range |
|---|---|---|---|
| NEB Ultra II | Ligation-based | 100pg - 100ng | 100nt-500nt |
| NexteraXT - NexteraFlex | Tn5 Transposase-based | 1ng - 300ng | adjustable minimum by SPRI cleanup |
NEB Ultra II
|
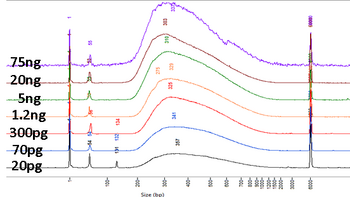
The BioMicro Center utilizes the NEB UltraII kit for standard library preparation. This kit is extremely robust and can make libraries from <1ng of input material. It is also amenable to miniaturization and is available as a high-throughput prep as well. NEB Ultra II is based on standard LM-PCR based chemistry. DNA fragments are repaired, A-tailed and ligated to adapters. The ligated products are then amplified by PCR to add indexes and the i5 and i7 anchors. Completed libraries can be size selected using SPRI beads or BluePippin. |
 |
Nextera Flex
|
Nextera FLEX uses bead-linked transposome chemistry for on-bead tagmentation. This provides steric inhibition which increases the size of the inserts in the library. At and above 100 ng DNA input, the beads are saturated and libraries are self-normalized. An automated version of Flex is available. For more information, please look at the High-Throuhgput DNA preparation page.
|
Nextera XT
|
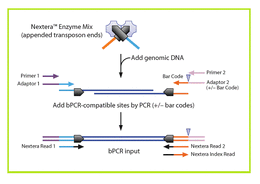
Nextera uses a modified Tn5 transposase to simultaneously fragment intact genomic DNA and tag it with Illumina adapters. A limited number of PCR steps are required to generate complete Illumina libraries. This very simple method makes this preparation very popular for automation and in vivo methods such as ATAC-Seq. Original Nextera requires 50 ng of input material and produces the most complex libraries. Products can be sized by Pippin and are suitable for all applications. NexteraXT uses less input material (1 ng) and less enzyme, making it less expensive, but only crude size fractionation using single-pass SPRI selection can be done. An automated version of Nextera XT is available. For more information, please look at the High-Throuhgput DNA preparation page. Nextera works well with amplicons as well as gDNA. However, because two hits are required per molecule to create productive libraries, the ratio of reagent to fragment must be altered significantly to produce good libraries and large inserts may not be possible. |
 |
