Barcodes
"A long time ago, in a building far, far away..." Randy designed some fancy barcodes for our BioBricks.
Here's the URL describing the original plan.
We've been building all our new ORF parts with barcodes for the last couple years.
More recently, with Heather's help, we ordered five sets of primers to finally test whether or not these barcodes work. Remember that the original purpose of the barcodes was to be able to provide a qualitative screen for whether or not a uncharacterized biological sample contains any BioBricks (or not). Nothing more fancy.
Meanwhile, there's also a project spun-up on vector barcodes.
The five different primer sets are...
Barcode_A_for: 5' TAG TGC TAG TGT AGA TCA C 3'
Barcode_A_rev: 5’ TAC ACT AGC ACT ATC AGA G 3'
Barcode_C_for: 5' TAG TGC TAG TGT AGA TCC C 3'
Barcode_C_rev: 5' TAC ACT AGC ACT ATC AGC G 3'
Barcode_G_for: 5' TAG TGC TAG TGT AGA TCG C 3'
Barcode_G_rev: 5' TAC ACT AGC ACT ATC AGG G 3'
Barcode_T_for: 5' TAG TGC TAG TGT AGA TCT C 3'
Barcode_T_rev: 5' TAC ACT AGC ACT ATC AGT G 3'
Barcode_Internal_for: 5' CTG ATA GTG CTA GTG TAG ATC 3'
Barcode_Internal_rev: 5' GAT CTA CAC TAG CAC TAT CAG 3'
Today, with Jen's help, we both ran six PCR reactions (each single primer pair plus all four base-specific primers in a single mix). We used four templates: purified synchronator DNA, MC4100 cells containing synchronator plasmid, MC4100 cells (not containing any BioBricks), water. The gel image is below.
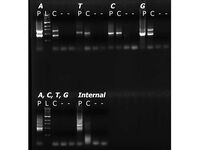
Going forward, based on conversations with Kathleen, it seems like we might want to consider improvements to our barcodes such as:
- using a slightly longer barcode with offset primers that can't form dimers
- using a barcode/primer combination that's got a slightly higher annealing temperature
- embed the parity check in a region that doesn't influence the primer (??)
- use primers that have a extra 5' section that is not part encoded barcode. this would let us increase the annealing temperature during the PCR reaction and help to improve specificity. importantly, any extra 5' section should not have homology to natural DNA sequence
Update: I just spoke with Randy about barcodes some. Randy reminded me that a number of folks (e.g., Pam Silver's lab) are working to adapt BioBricks for protein engineering, and that Austin has advanced BioBricks assembly schemes that are scarless. The short and the long of all this work is that we might consider moving from ORF-based barcodes to system specific barcodes. I.e., we could have a single BarCode part that all BioBrick systems include. The features of such a move would be:
- less risk of disrupting function,
- reduced or no risk of repeat-based homology deletions or other changes,
- a system-independent signature that could be made to work once, and wouldn't suffer from system-system variation.
- might be more compatible with protein engineering projects
Issue: Barry asks about whether or not sequencing will improve so fast, that a PCR-based detection scheme will be irrelevent by the time BioBricks are widely distributed.
- I don't think that improvement in sequencing technology will affect one of the original motivations for barcodes: Drew drinks a glass of water and his hair becomes pink. He would like to know whether this is his birthday present from Randy. Without barcodes, even with super-duper fast sequencing technology, it would require sequencing everything in the water. If Randy wishes to remain anonymous, all he should do is add a bunch of random DNA. As long as sequencing requires some positive amount of resources and time, it will always be cheaper and easier to thwart sequencing detection by adding random DNA. To thwart barcodes would involve a whole lot of genetic editing which is too much work, even for a birthday present for Drew. Austin 15:33, 11 Jun 2005 (EDT)
- There are two assumptions here. First, the builder wants their system to be found if it gets out. In this case, a bar code system might help. The advantages are described by Austin above. However,we may only need something as simple as putting a sequence into all deployed systems. The nice thing about the bar codes on the end of all coding sequences is that there is reduncancy. I don't buy that we will be able to deconvolute these systems by a PCR signature (but that I guess is up for debate). Anyways, point being, If we find the bar code, we sequence the whole region, and have somewhere to possibly start. However, if Randy were malicious there is a second assumption: Synthesis technologies will be impeded enough that potential malicious hackers will have no choice but to build their systems with bar codes, and were too lazy even to mutate this bar codes beyond recognition. I think this is a weak assumption to ride on. I think we should focus our attention on creating technologies for the first scenario of detecting accidental release. In this case, there may be less intrusive ways to mark the system. Finally, we should consider if our ability to make something dangerous is increasing much more slowly than our ability to sequence everything, as I think is occuring. In this case, we should rethink whether or not we should be barcoding anything. --Sri Kosuri 13:01, 28 Jul 2005 (EDT)
- UK Patent on Barcodes, with an eye on tagging GM stuff