User:Eleni N. Kalivas/Notebook/CHEM-571/2013/09/04
OverviewDue to time constraints we were not able to run a UV-vis for the inosine spectra. The objective for today includes analyzing yesterdays data in order to check the spectra and calibration curves and make any necessary corrections, and to also do the same for the inosine solutions. AnalysisAfter looking over the data from yesterday there were multiple problems that needed correcting. First we noticed that our 0.5M adenosine was the exact same data as the water that was run as a blank. This lead us to believe that the data was saved incorrectly and that some of the dilutions may have just been off. We re-ran the 0.5M adenosine sample and got comparatively valid data. We then noticed that our 1.0M adenosine peaked below the 0.5M which indicated a clear error. Therefore we re-made the 1.0M adenosine and received much better data. Our last observation was that the 3.0M adenosine was too far off when analyzing the calibration curve, so we also re-made the 3.0M adenosine dilution and re-ran it and received much better data.
InosineOnce the Adenosine samples had been run and the spectra and the calibration curve had been corrected, the inosine samples were now being run. After running all the data the spectra was plotted and the calibration curve was made. Analysis of the calibration curve concluded.
Group Calibration Data
the left is the standard deviation table of Adenosine, while that on the right is the standard deviation table of the Inosine data
the left is for Adenosine and the right is for the Inosine data
the left is for Adenosine and the right is for Inosine
Adenosine: Since Qc=0.64, the third and fourth points can be discarded with 90% confidence as outliers
Inosine: since Qc=0.64 the first four data points listed in the table can be discarded with 90% confidence as outliers
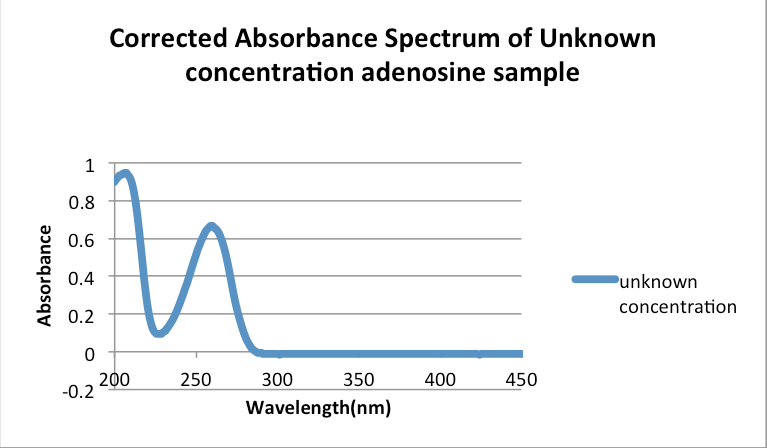
since Gc=1.67 the first three values, of 0.558, 0.442 and 0.406, listed on the table can be discarded with 95% confidence as outliers Identifying an unknown Adenosine sample
| |