20.309:Homeworks/Homework 3
General advice:
- There is no lab work required this week, but read the fluorescence microscopy lab manual to prepare for next week.
- Feel free to work on this homework set on your own computer. (You will need MatLAB plus the image processing toolbox.)
- If you would like some help, instructors and TAs will be available in the lab. In particular, TAs will be available on Tuesday from 3:30-5:30, Wednesday from 3:00-6:00, Thursday from 1:00-6:00, and Friday 1:00-5:00. The lab will be open regular hours.
- All the image files you will use in this homework can be found in the students folder of the course locker. Do not copy the low resolution images from this page.
- The images are of different sizes, file formats, and aspect ratios. All are grayscale. The techniques you will use to solve the problems on this homework generalize to multiple color channels, such as RGB.
Some useful commands:
imreadloads an image from a file. The syntax is:A = imread('filename'). The image data from the file is stored in matrix A.imshowdisplays the data of an image file in a matlab figure window. You use theimshowwith or without loading the image into the matlab workspace.imwritewrites an image to a file. You can specify a format. The basic syntax isimwrite(A,'filename').aviinforeturns the header information about a movie file in .avi formataviread('filename', 1)reads one frame of the avifile into a Matlab movie structure. If f is the movie structure, you can assign the image data of that frame to a matrix A using the command A = f.cdata.
Also:
- Pay attention to the numerical data type of these images when matlab imports them.
- You may need to convert between types.
- Use the workspace panel in the Matlab main window or the
whoandwhoscommands to get details on the size and type variables - Explore the rest of the commands in the image proessing toolbox in the Matlab help browser. You might want to take a look at: fft, fftshift, imhist, histeq, imadjust, conv, conv2, for example.
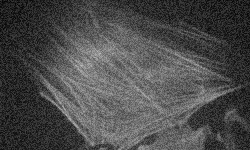
Problem 1:correcting poor contrast
Optimize the contrast for visualization of the microscope image on the left below, taken at too low a light level.
 |
 |
| dark actin.jpg | Contrast enhanced version |
Problem 2:filtering image data
Create a 3by3 and a 5by5 low pass filter. Apply these filters to remove noise from the cell image on the right above (noisy actin.tif). Can you make a 7by7 filter also?
Problem 3:impulse noise
Generating a long integration image using a CCD camera sometimes produces large intensity image spiky_actin1.tif. Can you develop a different method to remove the spikes if you have two pictures taken one after the other?

|

|
| spiky_actin1.tif | spiky_actin2.tif |
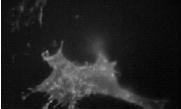
Problem 4:segmentation
Perform a segmentation to isolate the region corresponding to the actin stress fibers in the image actin_f.tif. Also perform a segmentation to isolate the region corresponding to clustered actin in a cell with a chemically disrupted cytoskeleton actin_treated.tif. The output of both tasks should be binary images corresponding to the desired regions.
 |
 |
| actin_f.tif | actin_treated.tif |
Problem 5:calculating magnification
You have two images of a "test pattern" consisting of light and dark line pairs with a spacing of 18 line-pairs per millimeter. This test pattern was imaged with a home-built microscope at two different magnifications. Calculate the magnification difference between these two images.
 |
 |
| HiMag.bmp | LowMag.bmp |
Problem 6:removing periodic noise
Periodic noise often occurs in biological imaging -- high noise.tif (below, left) & low noise.tif(below, center). Can you remove the noise if you know the (normalized) noise characteristics (noise.tif - below, right)?

|

|

|
| high noise.tif | low noise.tif | noise.tif |
Consider a case in which you do not know the noise characteristics. Can you clean up the image below? (For bonus credit, feel free to work on any AFM images of your own that you'd like to similarly clean up.)
 |
| ecoli noisy.jpg |
Problem 7:particle tracking
You are given a movie file (beadtest.avi) of a fluorescent bead in the cytoskeleton of the cell. Choose any bead in the image. Can you find the centroid of the bead as a function of frame number?