Biomod/2011/TeamJapan/Tokyo/Project/UV switch
<html> <!--<head>--> <!-- We refer to the wiki of team iGEM Heidlberg 2009. -->
<style type="text/css">
/* ====================
主に全体に関わるCSS
==================== */
body {behavior: url(http://www.xs4all.nl/~peterned/htc/csshover3-source.htc);}
.clear {clear:both;}
body.mediawiki {
font-size: 14px;
background-color:#707070; background-position: center center; background-attachment: fixed; background-repeat: no-repeat; font-family: Calibri, Verdana, helvetica, sans-serif; }
h1 {
padding: 0px 20px 5px 20px;
font-size: 34px;
font-weight: bold;
}
h2 {
padding: 20px 20px 5px 20px;
font-size: 25px;
color: #0083eb;
text-decoration: none;
font-weight: bold;}
border: none;
h2 a {
color: #eb8300;
}
h3 {
padding: 20px 20px 5px 20px;
font-size: 20px;
color: #000;
font-decoration: none;
font-weight: bold;
}
h1.firstHeading {
display: none;
}
p { text-align: justify; } a:link { color: #00a5ea; text-decoration: none } a:visited { color:#00a5ea; text-decoration: none } a:hover {
color: #eb8300;
text-decoration: none } a:active { color:#f29400; text-decoration: none } #bodyContent {
width: 970px;
margin: 0px 0px 0px;
background-color:#ffffff;
border-width: 0px 1px 0px 1px;
border-color: #000000;
}
#content {
padding-left: 0px; width: 970px;
}
table#team_members {
text-align: justify;
border: 0;
}
table#team_members h2, table#team_members h3 {
clear: both;
}
#content * a:hover {
text-decoration:none;
}
#main_wrapper {
position:absolute;
left:0px;
top:20px;
margin-top: 0; width: 969px; height: 221px; align: center; border-style: solid;
border-color: white;
} /* ====================
メニューの画像を変更できる部分 ==================== */
#header {
position:relative;
left:0px;
top:0px;
margin-top: 0; width: 969px; height: 221px; align: left; background-color: #FFFFFF;
background-image: url(http://openwetware.org/images/e/e0/NEW_header.jpg);
} /* ====================
以下、特殊なclassに適用される ==================== */
#navigation { position:absolute;
left:18px; top:155px; width:1200px; height:69px;
z-index:100;
background-color: transparent;
float: left;
color: #0000FF; } #super_main_wrapper {
position:absolute;
left:0px;
top:227px;
width: 975px; align: center; background-color: #ede8e2;
heigth: auto;
}
#SubWrapper {
width: 645px;
padding: 0px;
border-left:4px solid #ede8e2;
float: left;
margin-top: 0px;
background-color: #ede8e2;
}
#SubWrapper * p, #SubWrapper p {
padding: 0 20px;
text-align: justify;
font-size: 12px;
}
#SubWrapper * h3, #SubWrapper h3 {
padding-top: 10px;
font-size: 18px;
}
#news {
width: 322px;
margin-top: 0px;
float: left;
background-color: #d8d5d0;
border-right:4px solid #ede8e2;
}
#news p {
padding: 0 20px 20px 20px;
text-align: justify;
font-size: 12px;
}
#news h3 {
padding: 10px 20px;
font-size: 18px;
}
#mission_box {
width:650px;
float: left;
}
#team_box, #heartbeat_box, #notebook_box, #parts_box, #gallery_box, #sponsors_box, .boxy {
width:215px;
float: left;
padding: 10px 0 0 0;
}
div.tleft {
border-width: 0px;
margin:0;
padding:0;
border-color:transparent;
}
/* ====================
ここからプルダウン周辺のデザイン ==================== */
- menu * {
margin: 0; padding: 0; }
- menu {
behavior: url(http://www.xs4all.nl/~peterned/htc/csshover3-source.htc); <!-- fuer ie6 --> font-family: calibri, verdana, sans-serif;
font-color: #ffffff;
font-size: 19px; background-color: transparent; float:left; padding: 12px 0 0 0; }
- menu ul {
float: left; list-style: none; }
- menu ul li {
background-color:transparent;
position:relative;
float:left; list-style: none; padding: 10px 20px 0 0;
font-weight: bold;
width: auto;
}
- menu a {
color: #FFFFFF; display: inline; text-decoration: none; }
- menu a:visited {
color:#FFFFFF; text-decoration: none }
- menu a:hover {
color: #00a5ea; }
- menu ul li ul {
display: none; position: absolute; left: 0px;
width: 155px;
heigth: 1%;
font-size: 19px; opacity: 0.8; list-style: none;
top: 30px;
padding-top: 20px;
z-index:500;
}
- menu ul li:hover ul {
display: inline;
background-position: bottom;
}
- menu ul li ul li {
width: 100%; list-style: none;
background-color: #000;
margin: -1px;
padding: 0px 0 0 5px;
display: inline;
}
</style> <!-- </head> -->
<!-- ############################# BODY!!!!! #################################### -->
<!-- <body> --> <!--[if IE]> <style type="text/css">
- SubWrapper {width: 645px;}
- news {width: 317px;}
- super_main_wrapper {position:static;}
- navigation {left: 15px;}
- menu ul li ul {left: -2px; top: 22px;}
</style> <![endif]--> <div id="main_wrapper">
<div id="header">
<div id="navigation"> <div id="menu" style="position:static"> <ul> <li><a class="aMain" href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo">Home</a></li> <li><a class="aTeam" href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Team/Students">Team</a></li> <li><a class="aProject" href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project">Project</a> <!-- <ul> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project">Overview</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/introduction">Introduction</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Model">Model</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Devices">Devices</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Modes">Modes</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Results">Results</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Achievements">Achievements</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Future_works">Future works</a></li> </ul> --> <li><font color="#ffffff">Results</font> <ul> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Results">Experiments</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Simulations">Simulations</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Achievements/DNA_Devices">DNA Design</a></li> </ul></li> <!-- <li><a class="Simulation" href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Simulations">Simulations</a></li> <li><a class="DNA design" href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Achievements/DNA_Devices">DNA Designs</a></li> --> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Achievements">Achievements</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Project/Future_works">Future works</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Notebook/Protocols">Protocols</a></li> <li><a href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Notebook/Lab.notebook">Notes</a></li> <li><a class="aNotebook" href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Sponsors/">Sponsors</a></li> <li><a class="aSitemap" href="http://openwetware.org/wiki/Biomod/2011/TeamJapan/Tokyo/Sitemap">Sitemap</a></li>
</ul> </div><!-- end drop menu --> </div> <!-- end navigation --> </div> </div> <!-- For IE 6 --> <script type="text/javascript">
/***********************************************
- CSS Vertical List Menu- by JavaScript Kit (www.javascriptkit.com)
- Menu interface credits: http://www.dynamicdrive.com/style/csslibrary/item/glossy-vertical-menu/
- This notice must stay intact for usage
- Visit JavaScript Kit at http://www.javascriptkit.com/ for this script and 100s more
- /
var menuids=new Array("verticalmenu") //Enter id(s) of UL menus, separated by commas var submenuoffset=-2 //Offset of submenus from main menu. Default is -2 pixels.
function createcssmenu(){ for (var i=0; i<menuids.length; i++){
var ultags=document.getElementById(menuids[i]).getElementsByTagName("ul")
for (var t=0; t<ultags.length; t++){
var spanref=document.createElement("span")
spanref.className="arrowdiv" spanref.innerHTML=" " ultags[t].parentNode.getElementsByTagName("a")[0].appendChild(spanref)
ultags[t].parentNode.onmouseover=function(){
this.getElementsByTagName("ul")[0].style.left=this.parentNode.offsetWidth+submenuoffset+"px"
this.getElementsByTagName("ul")[0].style.display="block"
}
ultags[t].parentNode.onmouseout=function(){
this.getElementsByTagName("ul")[0].style.display="none"
}
}
}
}
if (window.addEventListener)
window.addEventListener("load", createcssmenu, false)
else if (window.attachEvent)
window.attachEvent("onload", createcssmenu)
</script>
<!-- </body> -->
</html>
DNA devices
Introduction
- We tried to three modes of DNA ciliate. Especially, to achieve two mode of DNA ciliate: “track walking mode” and “light-irradiated-gathering mode”, we designed two systems by DNAs and five types of DNA arrangements.
- Five types of DNA are followings. First is “deoxyribozyme” which is attached to DNA ciliate body. Second is “substrate” which is attached to a gloass plate as a scaffold of the track walking mode. Third is “UV-switching DNA” which is used for UV-switching system. Forth is “blocking DNA” which is also used for UV-switching system. Fifth is “complementary strand for deoxyribozyme” Especially, the third DNA, “UV-switching DNA” is the DNA strand which we designed by ourselves.
- We could check that all these five types of DNA strand work as we expected. This is worthy of special mention.
- In this page, we explain two systems: “track walking mode” and “UV-switching mode”, and explain five types of DNA strands which we designed. We write the results of checking DNA’s work by experimentation.
Design of DNA-DNA interaction mechanisms
Deoxyribozyme-substrate reaction for track walking mode

- Substrate and deoxyribozyme are used in this system. First, deoxyribozyme hybridizes with substrate. Second, deoxyribozyme cleaves substrate at ribose adenine and make two products. Third, the shouter product separates and the longer product:cleaved substrate hybridizes with deoxyribozyme. Forth, the hybridization becomes fragile and deoxyribozyme hybridizes with neighbor substrate. DNA ciliate repeats this reaction and move directly.
UV-switching system for light-irradiated gathering mode
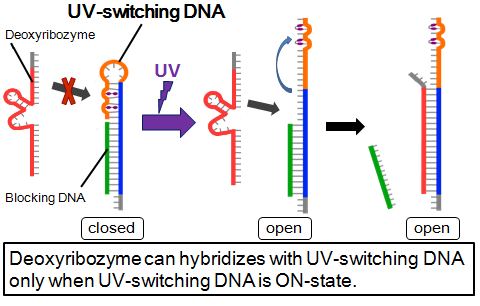
- Deoxyribozyme, blocking DNA, and UV-switching DNA are used in this system. UV-switching DNA has a stem-loop which contains two azobenzenes. By spotting UV, azobenzenes are isomerized and this loop becomes open. This opened loop has a complementary part for deoxyribozyme. This UV-switching DNA is complimentary for blocking DNA.
- In no UV, UV-switching DNA hybridizes with blocking DNA. This structure is OFF state and DNA ciliate’s deoxyribozyme doesn’t hybridize. By spotting UV, UV-switching DNA’s loop becomes open and branch migration starts, so DNA ciliate’s deoxyribozyme become trapped by UV-switching-DNA. This structure is ON state
- We used the way to use UV-switching DNA which is one DNA and has a loop, but there is the way to use two DNAs and insert azobenzens into the mate. Concretely, it is the way to insert azobenzens into the complementary DNA for substrate. By spotting UV, the complementary DNA is released and deoxyribozyme hybridizes with substrate. On the other hand, substrate can’t be inserted azobenzens because azobenzenes which are spotted UV inhibit hybridizing, so azobenzes inserted substrate can’t hybridize with not only complimentary DNA for azobenzenes inserted substrate but also deoxyribozyme.
- Thinking about UV-switching system in our project, it is an important thing that substrate is attached on a glass plate. Various reactions about DNA ciliates happen in solution, so the probability of molecules’ approaching to molecules on a glass plate is much smaller than the probability of molecules’ approaching to molecules in solution.
- Accordingly, to happen hybridizing and nurturing about substrate and its complementary DNA speedily, it’s necessary to heighten the probability of approaching. As a result, many complementary DNA for substrate doesn’t hybridize with substrate. The complementary DNA is very expensive DNA which azobenzenes are inserted, so the cost performance is very wrong.
- Furthermore, our system uses UV-switching DNA which has both azobenzenes area and its complementary area. Thanks to these areas, switching two areas are always close and in one DNA strand, so the UV-witching is smooth as making and collapsing a stem.
- In conclusion, comparing with the traditional system, our system is very reasonable and speedily for UV-switching.
Sequence Design
[Deoxyribozyme]
[Substrate]
[UV-switching DNA]
 |
 |
- 5' -(NH2)-TTTTTT TTTTCACTATTTCGACCGGCTCGGAGAAGAG TTTTT CT X CT X TC-3' (X means azobenzene. )
- Size: 48bases + 2azobenzenes
- UV-switching DNA is used for the scaffold in light-irradiated gathering mode. We designed this DNA by ourselves. UV-switching DNA has a five bases’ loop (TTTTT) and there are two azobenzenes (X) in the one side of the stem (CTXCTXCT).
- By spotting UV, azobenzenes are isomerized (trans to cis), so the part which contains azobenzenes becomes hard to form double strand. It is known that UV-switching can be realized by using this principle. (ref)
- To achieve this switching, it is necessary to design the stem which forms the loop firmly in the room temperature and opens the loop by isomerizing of two azobenzenes. We didn’t find the precedent which succeeded in opening and closing at a single molecular by azobenzenes which are inserted into a stem, so the designing is very difficult. After trial and error, we designed to use “GAAGAG” and “CTXCTXCT” as the stem and “TTTTT” as the loop.
- The 7th to 37th bases from 5' end (TTTTCACTATTTCGACCGGCTCGGAGAAGAG) is a complete complementary part for the deoxyribozyme. In addition, the 7th to 31th bases from 5’ end (TTTTCACTATTTCGACCGGCTCGGA) are a complementary part for blocking DNA. Consequently, the 32th to 37th bases from 5’ end (GAAGAG) are a complementary part for deoxyriboyme and not a complementary part for blocking DNA, so branch migration is started from this part and blocking DNA is released. Moreover, these 6 bases are a part which makes stem, so this part is blocked when the loop is closed. By this structure, branch migration doesn’t happen when the loop is formed.
- We designed the first 6 bases from 5' end as a linker (TTTTTT). This is also designed not to make unexpected structures. As a result, we decide using this linker
- The 5' end is aminated to be fixed on a glass plate.
[Blocking DNA]
[Complementary DNA for deoxyribozyme]
Confirmation of DNA-DNA interaction mechanisms
Deoxyribozyme-substrate reaction
- Observation result is here.
- Urea electrophoresis result is equal to DNA ciliate body’s page (link)
UV-switching system
- No urea electrophoresis result is here.
- U…UV-switching-trap-DNA
- B…Blocking DNA
- D…Deoxyribozyme DNA
- Reaction solution…A 0.225uM and B 0.45uM and D 0.225uM
- All solutions are in 5x SSC (sodium citrate 75mM)
- From left, these bands mean followings.
- U 0.225uM and Mg(2+) 80mM
- B 0.45uM and Mg(2+) 80mM
- D 0.225uM and Mg(2+) 80mM
- U 0.225uM and B 0.45uM and Mg(2+) 80mM
- U 0.225uM and D 0.225uM and Mg(2+) 80mM (UV isn’t spotted)
- U 0.225uM and D 0.225uM and Mg(2+) 80mM (UV is spotted for 60 min.)
- Reaction solution (UV isn’t spotted)
- Reaction solution (UV is spotted for 15 min.)
- Reaction solution (UV is spotted for 60 min.)
- Reaction solution and Mg(2+) 80mM (UV isn’t spotted)
- Reaction solution and Mg(2+) 80mM (UV is spotted for 15 min.)
- Reaction solution and Mg(2+) 80mM (UV is spotted for 60 min.)
- The controls (lane 1 to 6) meanings are following. Lane 4 (U 0.225uM and B 0.45uM and Mg(2+) 80mM) means the bands when the loop is stable and hybridization U and B (band U-B). Lane 5 (U 0.225uM and D 0.225uM and Mg(2+) 80mM (UV isn’t spotted)) means the bands when the loop is open and hybridization U and D. Lane 6 (U 0.225uM and D 0.225uM and Mg(2+) 80mM (UV is spotted for 60 min.)) means the bands when the loop is open and spotted UV (band A-D).
- If the switching is successful, band U-B becomes vague and band U-D becomes strong after spotting UV.
Complementary part for deoxyribozyme
- Observation result is here.
Conclusions
Trap DNA
- From the result of observation, DNA ciliates are gathering on the trap DNA, on the other hand, there are few DNA ciliates on the area of no trap DNA. This observation means that trap DNA works as DNA ciliates’ stopper.
Substrate DNA
- From the result of electrophoresis, substrate DNA is cleaved by deoxyribozyme DNA when there are Zn(2+) ions. This electrophoresis result means substrate DAN can work as substrate for deoxyribozyme DNA.
UV-switching-trap-DNA
- From the result of electrophoresis, UV-switching doesn’t work in no Mg(2+) reaction solution because band A-B and A-D don’t change through UV. On the other hand, UV-switching does work in Mg(2+) reaction solution perfectly because band A-B fades out and band A-D appears thorough UV. This result means UV-switching-trap-DNA works in 5x SSC and Mg(2+) ions solutions.
Protocol
- Observation
- How to observe is here.
- Electrophoresis
- How to do electrophoresis is here.
- How to make samples is here.





