User:Gevorg
 |
| ||||||
Intro
I am currently a postdoctoral fellow in the DeGrado lab at University of Pennsylvania. I am currently working on problems in engineering intermembrane as well as water-soluble proteins and design of protein-small molecule interactions. Prior to this, I was a graduate student in the Keating lab at MIT (since the summer of 2003), followed by a short time as a postdoctoral associate in the same lab. In my graduate work I dealt with problems of protein design and protein interaction prediction, which is detailed in my PhD Thesis. I received a Bachelor's degree in Biochemistry and one in Computer Science from University of Maryland Baltimore County in 2002.
Recent Work
DrawCoil 1.0
Helical wheel diagrams come in very handy when one wants to do a quick-and-dirty (but often insightful) analysis of sequence/structure relationships in coiled-coil oligomers. Frustratingly, to this day I have not been able to find a program that builds helical wheel diagrams. So I decided to write one myself and make it available on the net, so that other people don't have to go through the pain of building helical wheel diagrams from scratch every time. Introducing DrawCoil 1.0. It is very simple to use (literally, just paste in the sequences and hit "submit"), but at the same time it is very highly configurable and you can control residue coloring, line widths, whether the bridges are shown and so on. Plus, new controllable features are easy to incorporate, so if there happen to be some commonly needed options, I'll try to put them in.

I defended!
Yay! Here is my PhD Thesis.

Computing van der Waals energies in the context of the rotamer approximation
This paper finally came out in "Early View" on the website of Proteins: Structure, Function and Bioinformatics. Hopefully, it will be in print within a month or so...

In many computational approaches to protein structure, the flexibility of amino-acid sidechains is represented via a finite set of rigid rotational isomers, known as rotamers. This representation is structurally justified (i.e. there is indeed a small set of conformations, which describe most of the flexibility of sidechains in proteins) and simplifies many of the aspects of the calculations. However, potential energies of protein conformations calculated using this so-called rotamer approximation are not necessarily in good agreement with the "true" potential energies due to the sensitivity of some energy terms to precise atomic location (see the schematic diagram below, where RCE and NCE represent the rotamer-based and true energy landscapes). The idea behind this study was to elucidate the extent to which this is a problem for such applications as computation protein design and structure prediction and to test the variety of simple "hacks" that are used in the field to address this problem.
Ultra-Fast Evaluation of Protein Energies Directly from Sequence
Our latest results on cluster expansion work was published in PLoS Computational Biology on 06/16/06. Click here for full access to the paper.

This paper describes how the theory of cluster expansion, originally developed to describe the energies of alloys, can be applied to generate a physical potential for proteins that is extremely fast to evaluate.
bZIP coiled-coil dimerization model
12/31/05. bZIP paper finally published: G. Grigoryan and A. E. Keating, "Structure-based Prediction of bZIP Partnering Specificity", J. Mol. Biol. (2006) 355, 1125-1142. PDF (MIT access only) Abstract
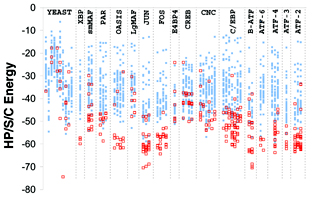
Gevorg Grigoryan and Amy E. Keating, "Structure-based prediction of bZIP partnering specificity", J. Mol. Biol., in press.
In general, our lab is interested in studying (experimentally and computationally) protein-protein interactions. In this project we looked at dimerization preferences among a family of eukaryotic transcription factors - basic region leucine zipper (bZIP) proteins. In an earlier study done in the lab1, protein microarrays had been used to experimentally characterize all pair-wise interactions among human bZIPs. In the present study, we used a computational structure prediction-based approach to explain the experimentally observed binding preferences. Our study presents a physically-relevant model that predicts results in good agreement with the experiment. We also outline certain successes and failures encountered along the way to developing the final model, which are likely relevant for many of the current structure-based approaches for prediction of interaction specificities. I will post the link as soon as the paper is published.
[1] John R. S. Newman and Amy E. Keating, "Comprehensive Identification of Human bZIP Interactions with Coiled-Coil Arrays", Science 300, 2097- 2101 (2003).
MIT TechTalk Article
10/26/05. MIT TechTalk published an article about our application of cluster expansion to protein design and the PRL paper. See here or here.
Fast protein sequence-energy mapping with Cluster Expansion
09/30/05. Our paper got published in Phys. Rev. Lett. Apparently, PRL has a super strict policy about reproducing papers in any format, so I can not post the PDF here, as previously promised. However, here is a link to the abstract. Our library has a subscription, so if you log in through Vera, you can get the PDF.

In collaboration with the Ceder Lab in the Department of Materials Science and Engineering at MIT, we have recently submitted a paper to Physics Review Letters (Fei Zhou, Gevorg Grigoryan, Amy Keating, Gerbrand Ceder, Dane Morgan, "Coarse graining protein energetics with cluster expansion"). It got accepted with some very positive reviews. Here are some excerpts:
- "This submitted paper is very original and could lead to important developments."
- "In other words, the authors might consider a somewhat "dumbed down" version of their paper (sorry!), and use it as a sort of introduction to a longer paper where mathematical developments are given in detail. In that way, readers in both disciplines may well look forward with great anticipation to the full treatment (to appear in PRB, say)... In summary, the authors' is an extremely interesting approach, perhaps the most wildly interdisciplinary that I have seen. It amply merits publication in PRL, but in my opinion requires reworking so that non-specialists (of both communities!) may appreciate the novelty and power of the proposed method."
- "Given the enormous interest in protein design and protein folding at the present time, the article is likely to find wide interest amongst the readership of the journal."
We are extremely happy that the paper got in with such great reviews. I shall post a PDF once it is published.
Contact Info
Department of Biochemistry and Biophysics
Stellar-Chance Building, Room 1015
422 Curie Blvd, Philadelphia, PA 19107
443-629-1227
gevorg@alum.mit.edu
gevorg@mail.med.upenn.edu
gevorg.grigoryan@gmail.com